Plots and images for papers¶
This notebook is used to generate various plots and images for publications and slide decks. It is not well-documented, but provides tips that show how to plot various things.
In [1]:
import numpy as np
import glob,os
%pylab inline --no-import-all
plt.rc('font', family='serif', serif='Times',size=15)
plt.rc('text', usetex=True)
plt.rc('xtick', labelsize=20)
plt.rc('xtick.major', size=10)
plt.rc('ytick.major', size=10)
plt.rc('ytick', labelsize=20)
plt.rc('axes', labelsize=20)
plt.rc('figure',titlesize=25)
plt.rcParams['image.origin'] = 'lower'
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['axes.linewidth'] = 2.
import logging as log
from crispy.tools.initLogger import getLogger
log = getLogger('main')
from crispy.tools.image import Image
Populating the interactive namespace from numpy and matplotlib
In [2]:
os.chdir('/Users/mrizzo/IFS/crispy/crispy/PISCES/')
from params import Params
par = Params()
par.hdr
0.7
In [2]:
cmap = 'RdBu'
cmap = 'plasma'
Constructing new colormap¶
In [3]:
import matplotlib as mpl
C = np.array([(69,6,90), (42,118,142), (73,193,109), (243,229,30), (255,255,255)]).T
# C = np.array([(255,255,255),(243,229,30),(73,193,109),(42,118,142),(69,6,90)]).T
from scipy.interpolate import interp1d
fitvector = [0.0,1.,2.,3,4.]
vector = np.arange(0.0,C.shape[1]-1,0.01)
Rinter = interp1d(fitvector,C[0,:])
R = Rinter(vector)
Ginter = interp1d(fitvector,C[1,:])
G = Ginter(vector)
Binter = interp1d(fitvector,C[2,:])
B = Binter(vector)
newC = np.zeros((3,len(vector)))
newC[0,:] = R
newC[1,:] = G
newC[2,:] = B
cm = mpl.colors.ListedColormap(newC.T/255.0)
cmap=cm
Flatfield¶
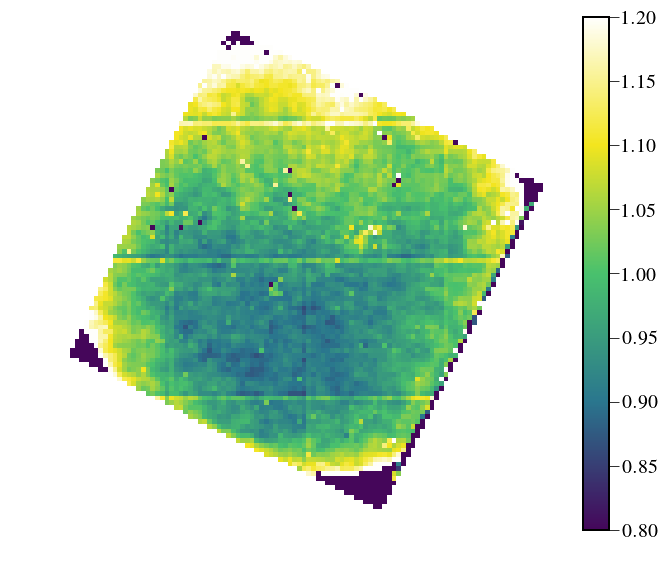
In [4]:
lenslet_flat = Image(par.wavecalDir+"/lenslet_flat.fits").data
lenslet_mask = Image(par.wavecalDir+"/lenslet_mask.fits").data
plt.figure(figsize=(10,10))
plt.imshow(lenslet_flat*lenslet_mask, cmap=cmap,vmin=0.8,vmax=1.2)
plt.colorbar(fraction=0.046, pad=0.04)
plt.savefig("/Users/mrizzo/Downloads/PISCESthroughput.png")
plt.axis('off')
plt.show()
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/Calibra_170425//lenslet_flat.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/Calibra_170425//lenslet_mask.fits
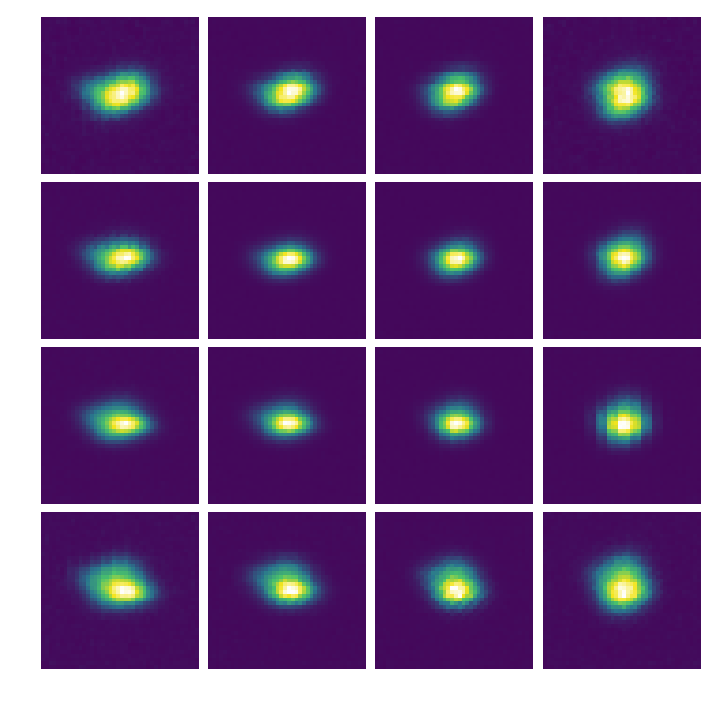
PSFLets¶
In [5]:
psflets = Image(par.wavecalDir+"/hires_psflets_lam715.fits").data
fig, axarr = plt.subplots(4,4,figsize=(12,12))
for i in range(psflets.shape[0]):
for j in range(psflets.shape[1]):
axarr[i,j].imshow(psflets[i,j],vmin=0.0,cmap=cmap)
axarr[i,j].axis('off')
plt.subplots_adjust(wspace=0.0,hspace=0.05)
plt.savefig(par.wavecalDir+"psflets.png")
plt.savefig("/Users/mrizzo/Downloads/Psflets.png")
plt.show()
crispy - INFO - Read data from HDU 0 of ..//ReferenceFiles/Calibra_170425//hires_psflets_lam715.fits
Extract cutout of microspectrum¶
In [6]:
from crispy.tools.locate_psflets import PSFLets
from crispy.tools.reduction import get_cutout
from crispy.unitTests import testCutout
lensnum = 0
subim = testCutout(par,par.codeRoot+'/Inputs/BB.fits',lensX=lensnum,lensY=lensnum)
polychrome = Image(par.wavecalDir+'/polychromeR70.fits').data
polyshape = polychrome.shape
fig, axarr = plt.subplots(1,12,figsize=(12,12))
print axarr.shape
axarr[0].imshow(subim.T,cmap=cmap)
axarr[0].axis('off')
for i in range(1,polyshape[0]+1):
axarr[i].imshow(testCutout(par,polychrome[i-1],lensX=lensnum,lensY=lensnum).T,cmap=cmap)
axarr[i].axis('off')
plt.subplots_adjust(wspace=0.1,hspace=0.05)
plt.savefig('/Users/mrizzo/Downloads/Microspectrum.png',dpi=200)
plt.show()
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
crispy - INFO - Read data from HDU 0 of ..//ReferenceFiles/Calibra_170425//polychromeR70.fits
(12,)
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
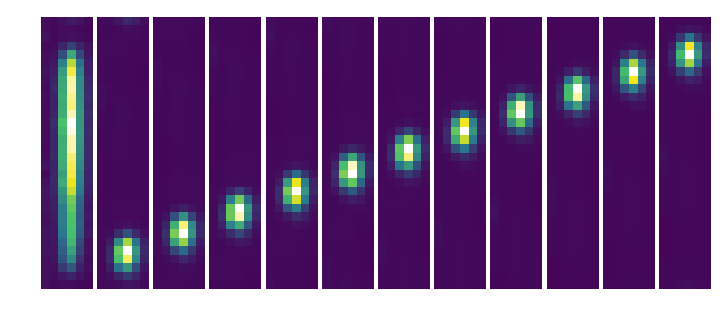
In [7]:
from crispy.tools.locate_psflets import PSFLets
from crispy.tools.reduction import get_cutout,calculateWaveList
from crispy.unitTests import testCutout,testFitCutout
lensnum = 0
lam_midpts,lam_endpts = calculateWaveList(par)
subim = testCutout(par,par.codeRoot+'/Inputs/BB.fits',lensX=lensnum,lensY=lensnum)
spectrum = testFitCutout(par,par.codeRoot+'/Inputs/BB.fits',lensX=lensnum,lensY=lensnum)
print "Spectrum",spectrum[0]
polychrome = Image(par.wavecalDir+'/polychromeR70.fits').data
polyshape = polychrome.shape
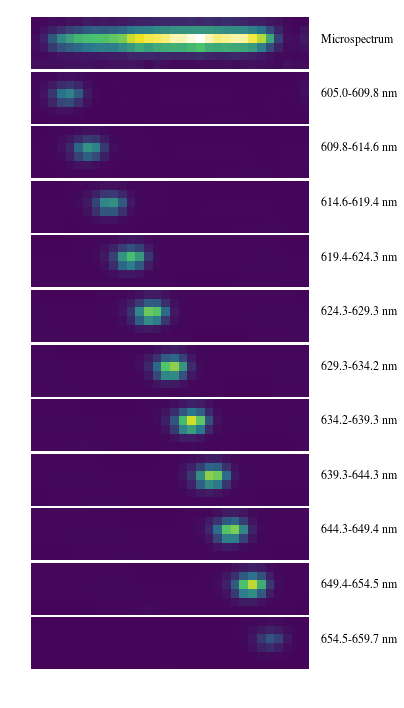
fig, axarr = plt.subplots(12,figsize=(12,12))
print axarr.shape
axarr[0].imshow(subim,cmap=cmap)
axarr[0].annotate('Microspectrum',xy=(33,2.5),annotation_clip=False,fontsize=12)
axarr[0].axis('off')
for i in range(1,polyshape[0]+1):
axarr[i].imshow(spectrum[0][i-1]*testCutout(par,polychrome[i-1],lensX=lensnum,lensY=lensnum),cmap=cmap,vmin=0.0,vmax=np.amax(subim))
axarr[i].annotate('%.1f-%.1f nm' % (lam_endpts[i-1],lam_endpts[i]),xy=(33,2.5),annotation_clip=False,fontsize=12)
axarr[i].axis('off')
plt.subplots_adjust(wspace=0.00,hspace=0.05)
plt.savefig('/Users/mrizzo/Downloads/MicrospectrumVertical.png',dpi=200)
plt.show()
crispy - INFO - Reduced cube will have 23 wavelength bins
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
Spectrum [ 258.86676159 298.12528886 290.42239038 403.19205797 462.7638815
476.37566957 530.84311221 458.47346772 419.55180325 454.847684
117.46899576]
crispy - INFO - Read data from HDU 0 of ..//ReferenceFiles/Calibra_170425//polychromeR70.fits
(12,)
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
Test fit cutout¶
In [20]:
from crispy.unitTests import testFitCutout
from crispy.IFS import reduceIFSMap
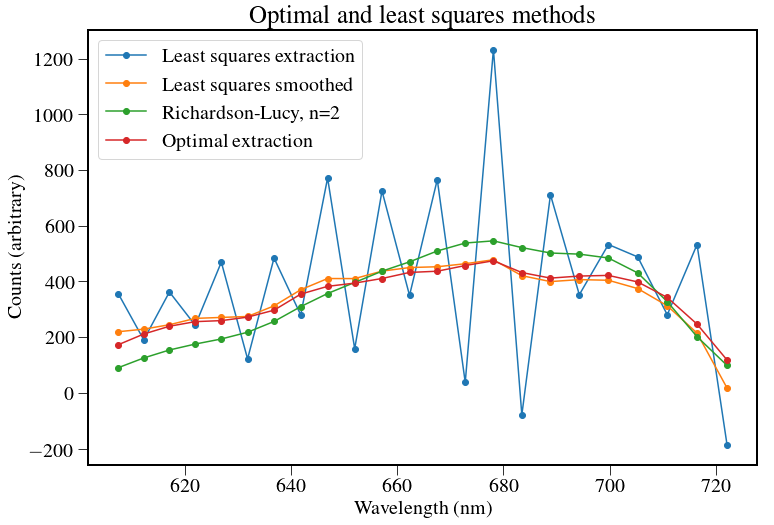
niter=2
spectral_cube = reduceIFSMap(par,par.codeRoot+'/Inputs/BB.fits',method='lstsq',smoothbad = False)
spectral_cube2 = reduceIFSMap(par,par.codeRoot+'/Inputs/BB.fits',method='lstsq_conv',smoothbad = False)
RL_cube = reduceIFSMap(par,par.codeRoot+'/Inputs/BB.fits',method='RL',smoothbad = False,niter=niter)
spectrum = spectral_cube.data[:,54,54]
spectrum2 = spectral_cube2.data[:,54,54]
spectrumRL = RL_cube.data[:,54,54]
optspect = reduceIFSMap(par,par.codeRoot+'/Inputs/BB.fits',method='optext')
print optspect.data[:,54,54]
plt.figure(figsize=(12,8))
lam_midpts,lam_endpts = calculateWaveList(par)
print lam_midpts
# plt.plot(lam_midpts,spectrum,linewidth=2,color='#1f77b4')
# plt.plot(lam_midpts,spectrum,'o-',color='#1f77b4')
plt.plot(lam_midpts,spectrum,'o-')
plt.plot(lam_midpts,spectrum2,'o-')
ratio = np.mean(spectrum2)/np.mean(spectrumRL)
plt.plot(lam_midpts,spectrumRL*ratio,'o-')
lam_midpts,lam_endpts = calculateWaveList(par,method="optext")
print lam_midpts
spect = optspect.data[:,54,54]
ratio = np.mean(spectrum2)/np.mean(spect)
print ratio
# plt.plot(lam_midpts,spect*ratio,'o-',color='#d62728')
plt.plot(lam_midpts,spect*ratio,'o-')
plt.tick_params(axis='both', which='major', labelsize=20)
plt.xlabel('Wavelength (nm)',fontsize=20)
plt.ylabel('Counts (arbitrary)',fontsize=20)
plt.title('Optimal and least squares methods',fontsize=25)
plt.legend(["Least squares extraction","Least squares smoothed","Richardson-Lucy, n=%d" % niter,"Optimal extraction"],fontsize=20)
plt.savefig('/Users/mrizzo/Downloads/OptimalVSlstsq.png',dpi=200)
plt.show()
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
crispy - INFO - Reduced cube will have 23 wavelength bins
crispy - INFO - Writing data to ..//SimResults/BB_red_lstsq_resid.fits
crispy - INFO - Writing data to ..//SimResults/BB_red_lstsq_model.fits
crispy - INFO - Elapsed time: 9.811288s
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
crispy - INFO - Reduced cube will have 23 wavelength bins
crispy - INFO - Writing data to ..//SimResults/BB_red_lstsq_conv_resid.fits
crispy - INFO - Writing data to ..//SimResults/BB_red_lstsq_conv_model.fits
crispy - INFO - Elapsed time: 86.092133s
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
crispy - INFO - Reduced cube will have 23 wavelength bins
crispy - INFO - Writing data to ..//SimResults/BB_red_RL_resid.fits
crispy - INFO - Writing data to ..//SimResults/BB_red_RL_model.fits
crispy - INFO - Elapsed time: 9.496640s
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
crispy - INFO - Reduced cube will have 23 wavelength bins
crispy - INFO - Elapsed time: 1.851557s
[ 966.72715824 1181.13275563 1336.82460971 1429.66716317 1449.88065036
1520.06275589 1659.48795903 1982.06303647 2137.83299707 2202.92063354
2295.18410426 2416.95325151 2441.46158831 2555.72005129 2651.14485437
2414.65131713 2303.03554857 2344.38337066 2357.46086189 2226.14746258
1914.69159057 1379.19412664 659.5318317 ]
crispy - INFO - Reduced cube will have 23 wavelength bins
[ 607.38448306 612.18168006 617.01676591 621.89003987 626.80180353
631.75236091 636.7420184 641.77108481 646.8398714 651.94869189
657.09786247 662.28770183 667.51853118 672.79067426 678.10445737
683.46020938 688.85826179 694.29894867 699.78260676 705.30957546
710.88019683 716.49481565 722.15377942]
crispy - INFO - Reduced cube will have 23 wavelength bins
[ 607.38448306 612.18168006 617.01676591 621.89003987 626.80180353
631.75236091 636.7420184 641.77108481 646.8398714 651.94869189
657.09786247 662.28770183 667.51853118 672.79067426 678.10445737
683.46020938 688.85826179 694.29894867 699.78260676 705.30957546
710.88019683 716.49481565 722.15377942]
0.178905387765
In [21]:
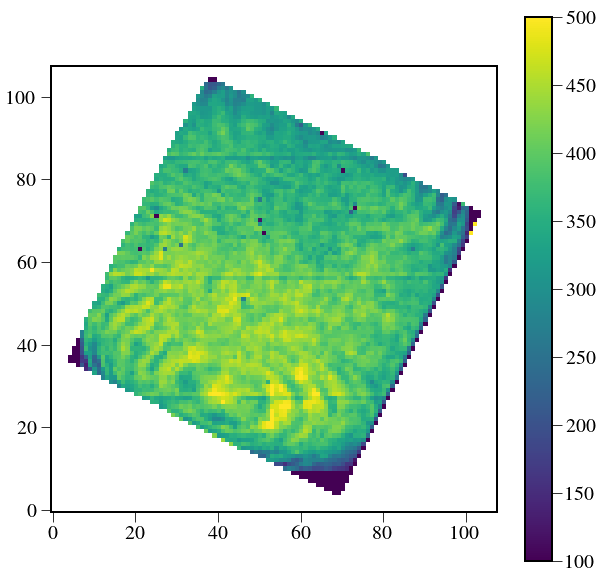
plt.figure(figsize=(10,10))
plt.imshow(spectral_cube2.data[9],vmin=100,vmax=500)
plt.colorbar()
Out[21]:
<matplotlib.colorbar.Colorbar at 0x18179bb790>
Optimal extraction¶
In [22]:
from crispy.tools.locate_psflets import PSFLets
from crispy.tools.reduction import get_cutout,calculateWaveList
from crispy.unitTests import testCutout,testFitCutout
import scipy
from scipy.special import erf
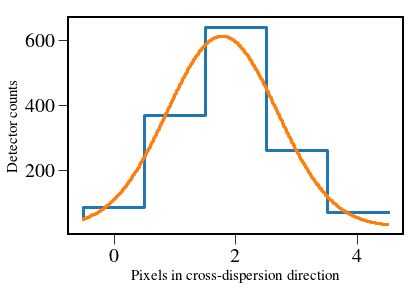
lensnum = 0
slicenum_list = [10]
for i in range(len(slicenum_list)):
slicenum=slicenum_list[i]
lam_midpts,lam_endpts = calculateWaveList(par)
subim = testCutout(par,par.codeRoot+'/Inputs/BB.fits',lensX=lensnum,lensY=lensnum)
plt.figure(figsize=(6,4))
x = np.arange(len(subim.T[slicenum]))-0.5
sig=2/2.35
plt.step(x,subim.T[slicenum],linewidth=3)
x2 = np.arange(0,len(subim.T[slicenum])-1,0.01)-0.5
val = np.mean(np.sum(x[:,np.newaxis]*subim,axis=0)/np.sum(subim,axis=0))
template = np.exp(-(x-val)**2/2./sig**2)/sig/np.sqrt(2.*np.pi)
template = (erf((x + 0.5 -val) / (np.sqrt(2) * sig)) - erf((x - 0.5-val) / (np.sqrt(2) * sig)))
val2 = val-0.5
template2 = (erf((x2 + 0.5 -val2) / (np.sqrt(2) * sig)) - erf((x2 - 0.5-val2) / (np.sqrt(2) * sig)))
b, a, _, _, _ = scipy.stats.linregress(template,subim.T[slicenum])
plt.step(x2,a+b*template2,linewidth=3)
plt.xlabel('Pixels in cross-dispersion direction',fontsize=15)
plt.ylabel('Detector counts',fontsize=15)
#plt.legend(["Pixel values in column","Gaussian matched filter"],fontsize=15)
plt.savefig('/Users/mrizzo/Downloads/Optimal_extraction.png',dpi=200)
plt.show()
crispy - INFO - Reduced cube will have 23 wavelength bins
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
[518, 524, 537, 569]
crispy - INFO - Writing data to ..//unitTestsOutputs/cutout.fits
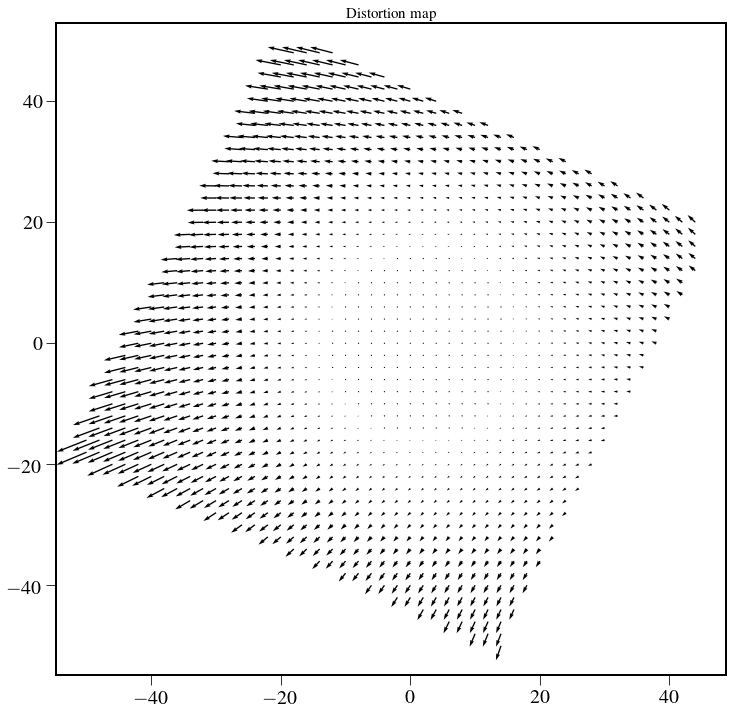
Plot distortion map¶
After thinking more about this, this is maybe not the correct way to calculate the distortion map…
In [23]:
from crispy.tools.locate_psflets import PSFLets
psftool = PSFLets()
lamlist = np.loadtxt(par.wavecalDir + "lamsol.dat")[:, 0]
allcoef = np.loadtxt(par.wavecalDir + "lamsol.dat")[:, 1:]
psftool.geninterparray(lamlist, allcoef)
# xindx = np.arange(-par.nlens/2, par.nlens/2)
# xindx, yindx = np.meshgrid(xindx, xindx)
xindx = np.arange(-50, 45)
yindx = np.arange(-50, 50)
lx = len(xindx)
ly = len(yindx)
xindx, yindx = np.meshgrid(xindx, yindx)
print lamlist[6]
allcoef_ideal = np.zeros(allcoef.shape)
indx = np.asarray([0, 1, 4, 10, 11, 14])
allcoef_ideal[:,indx] = allcoef[:,indx]
x,y = psftool.return_locations(lamlist[6], allcoef, yindx, xindx)
borderpix=4
good = (x > borderpix)*(x < 1024-borderpix)*(y > borderpix)*(y < 1024-borderpix)
psftool.geninterparray(lamlist, allcoef_ideal)
X,Y = psftool.return_locations(lamlist[6], allcoef_ideal, yindx, xindx)
U = x-X
V = y-Y
U[~good]= np.NaN
V[~good]= np.NaN
plt.figure(figsize=(12,12))
Q = plt.quiver(xindx[::2,::2], yindx[::2,::2], U[::2,::2], V[::2,::2],width=0.002,scale=700)
#plt.colorbar(fraction=0.046, pad=0.04)
plt.title('Distortion map',fontsize=15)
#qk = plt.quiverkey(Q, 0.9, 0.9, 2, r'$2 \frac{m}{s}$', labelpos='E',
# coordinates='figure')
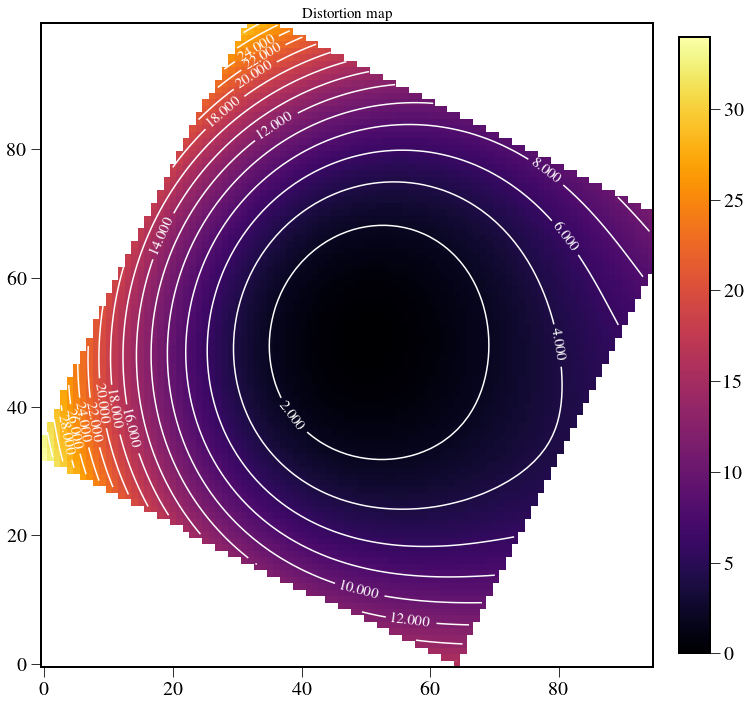
plt.figure(figsize=(12,12))
plt.imshow(np.sqrt(U**2+V**2),cmap=cmap)
plt.title('Distortion map',fontsize=15)
plt.colorbar(fraction=0.046, pad=0.04)
print np.median(xindx[0,:]),np.median(yindx[:,0])
CS = plt.contour(xindx-np.median(xindx[0,:])+lx//2,yindx-np.median(yindx[:,0])+ly//2,np.sqrt(U**2+V**2),20,colors='w')
plt.clabel(CS, fontsize=15, inline=1)
plt.show()
665.0
-3.0 -0.5
In [24]:
from crispy.tools.locate_psflets import PSFLets
psftool = PSFLets()
lamlist = np.loadtxt("/Users/mrizzo/IFS/charis-dep/code/calibrations/lowres/lamsol.dat")[:, 0]
allcoef = np.loadtxt("/Users/mrizzo/IFS/charis-dep/code/calibrations/lowres/lamsol.dat")[:, 1:]
print lamlist
import sys
psftool.geninterparray(lamlist, allcoef)
newcoefs = psftool.monochrome_coef(1550., lamlist, allcoef, order=3)
print newcoefs
[ 1096.63315843 1107.6545049 1118.78661775 1130.03061019 1141.38760663
1152.85874278 1164.44516577 1176.14803425 1187.96851851 1199.90780061
1211.96707449 1224.14754609 1236.45043347 1248.87696691 1261.4283891
1274.10595517 1286.91093291 1299.8446028 1312.90825825 1326.10320561
1339.43076439 1352.89226737 1366.48906071 1380.22250409 1394.09397087
1408.1048482 1422.2565372 1436.55045304 1450.98802511 1465.5706972
1480.29992758 1495.17718919 1510.20396976 1525.38177199 1540.71211367
1556.19652784 1571.83656296 1587.63378304 1603.58976783 1619.70611293
1635.98443 1652.42634686 1669.03350774 1685.80757337 1702.75022115
1719.86314538 1737.14805735 1754.60668558 1772.24077593 1790.05209184
1808.04241446 1826.21354282 1844.56729405 1863.10550356 1881.83002516
1900.74273134 1919.84551337 1939.14028156 1958.62896539 1978.31351375
1998.1958951 2018.27809772 2038.56212982 2059.05001984 2079.74381657
2100.64558942 2121.75742858 2143.08144525 2164.61977185 2186.37456223
2208.34799189 2230.54225819 2252.95958057 2275.60220079 2298.47238312
2321.57241461 2344.90460528 2368.47128836 2392.27482054 2416.31758219
2440.60197762]
[ 1.02809860e+03 -6.91921134e+00 -3.48952882e-04 5.25813509e-07
-1.36282008e+01 -5.30316402e-04 1.14037045e-06 3.55879515e-04
1.11823024e-06 9.28878964e-07 1.02061218e+03 1.33163261e+01
6.95924235e-04 -9.96713088e-07 -6.76149699e+00 -6.20708415e-04
1.61432926e-07 2.71700650e-04 -9.64295074e-07 3.38415501e-07]
Generic SPIE images¶
In [25]:
cmap='inferno'
In [26]:
%pylab inline --no-import-all
from crispy.tools.reduction import calculateWaveList
import matplotlib as mpl
from matplotlib.colors import LogNorm
lam_midpts,lam_endpts = calculateWaveList(par,method='optext')
print lam_midpts
print 660.*np.linspace(1.-0.18/2,1.+0.18/2,45)
cube = Image(filename='/Users/mrizzo/IFS/OS5/with_lowfc/os5_spc_031.fits').data
print cube.shape
plt.figure()
plt.imshow(np.sqrt(cube[15,70:-70,40:-40]),cmap=cmap)
print cube[15,70:-70,40:-40].shape
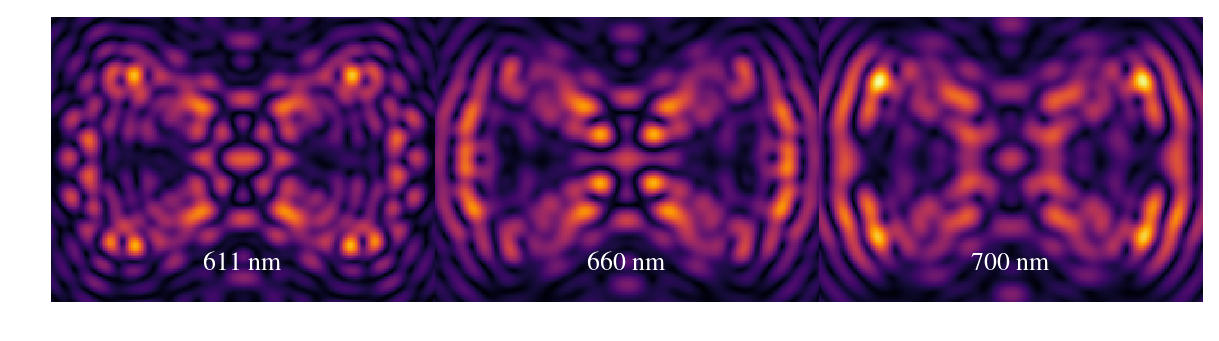
h,w = cube[15,70:-70,40:-40].shape
outarray = np.zeros((h,w*3))
outarray[:,:w] = cube[4,70:-70,40:-40]
outarray[:,w:w*2] = cube[22,70:-70,40:-40]
outarray[:,w*2:] = cube[-8,70:-70,40:-40]
plt.figure(figsize=(16,4))
plt.imshow(np.sqrt(outarray),vmin=1e-14,cmap=cmap)
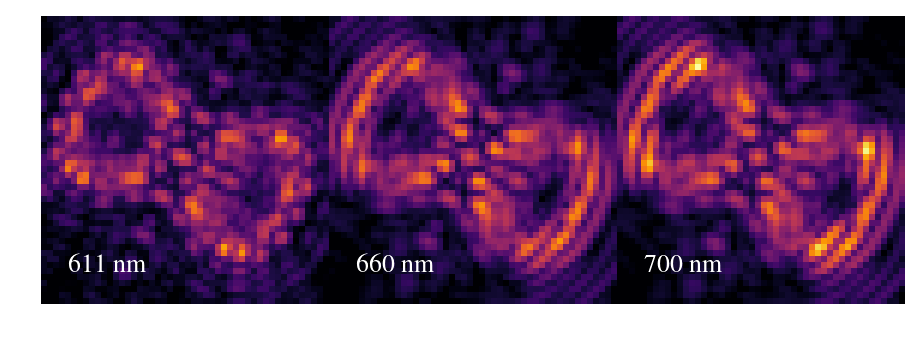
plt.axis('off')
plt.annotate('611 nm',xy=(w/2.-25,20),color='white',fontsize=26)
plt.annotate('660 nm',xy=(w+w/2.-25,20),color='white',fontsize=26)
plt.annotate('700 nm',xy=(2*w+w/2.-25,20),color='white',fontsize=26)
print np.amax(outarray)
plt.tight_layout()
plt.subplots_adjust(left=0.0, right=1, top=1, bottom=0)
plt.savefig("/Users/mrizzo/Downloads/InputSlices.png",dpi=200)
Populating the interactive namespace from numpy and matplotlib
crispy - INFO - Reduced cube will have 23 wavelength bins
[ 607.38448306 612.18168006 617.01676591 621.89003987 626.80180353
631.75236091 636.7420184 641.77108481 646.8398714 651.94869189
657.09786247 662.28770183 667.51853118 672.79067426 678.10445737
683.46020938 688.85826179 694.29894867 699.78260676 705.30957546
710.88019683 716.49481565 722.15377942]
[ 600.6 603.3 606. 608.7 611.4 614.1 616.8 619.5 622.2 624.9
627.6 630.3 633. 635.7 638.4 641.1 643.8 646.5 649.2 651.9
654.6 657.3 660. 662.7 665.4 668.1 670.8 673.5 676.2 678.9
681.6 684.3 687. 689.7 692.4 695.1 697.8 700.5 703.2 705.9
708.6 711.3 714. 716.7 719.4]
crispy - INFO - Read data from HDU 0 of /Users/mrizzo/IFS/OS5/with_lowfc/os5_spc_031.fits
(45, 315, 315)
(175, 235)
3.35319633686e-13
In [27]:
cmap='inferno'
IFS_detector = Image(filename='/Users/mrizzo/IFS/OS5_SIM_2.0_noiseless/average/average_target_star_detector.fits').data
import matplotlib.colors as colors
plt.figure(figsize=(9.0,6))
img = IFS_detector[300:-300,200:-200]
print np.amin(img)
plt.imshow(img,cmap=cmap,vmin=1e-5,norm=colors.PowerNorm(gamma=1./2.))
# plt.colorbar(fraction=0.046, pad=0.04)
plt.axis('off')
plt.tight_layout()
plt.subplots_adjust(left=0.0, right=1, top=1, bottom=0)
plt.savefig("/Users/mrizzo/Downloads/DetectorMap.png",dpi=200)
crispy - INFO - Read data from HDU 1 of /Users/mrizzo/IFS/OS5_SIM_2.0_noiseless/average/average_target_star_detector.fits
2.31331e-17
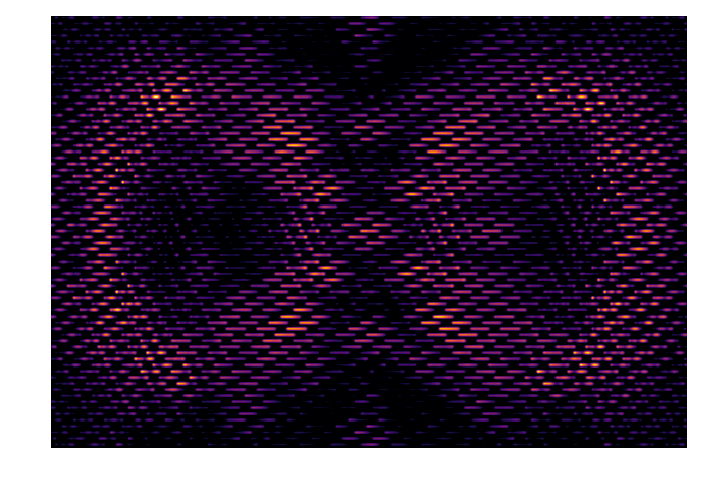
In [28]:
IFS_detector = Image(filename='/Users/mrizzo/IFS/OS5_SIM_2.0_t1000/average_47Umac/average_target_star_detector.fits').data
import matplotlib.colors as colors
plt.figure(figsize=(10,6))
img = IFS_detector[300:-300,200:-200]
print np.amin(img)
plt.imshow(img,cmap='inferno',vmin=1e-5,vmax=1e-3)
plt.colorbar(fraction=0.046, pad=0.04)
plt.axis('off')
plt.tight_layout()
plt.subplots_adjust(left=0.0,bottom=0)
plt.savefig("/Users/mrizzo/Downloads/NoisyDetectorMap.png",dpi=200)
crispy - INFO - Read data from HDU 1 of /Users/mrizzo/IFS/OS5_SIM_2.0_t1000/average_47Umac/average_target_star_detector.fits
3.67094e-05
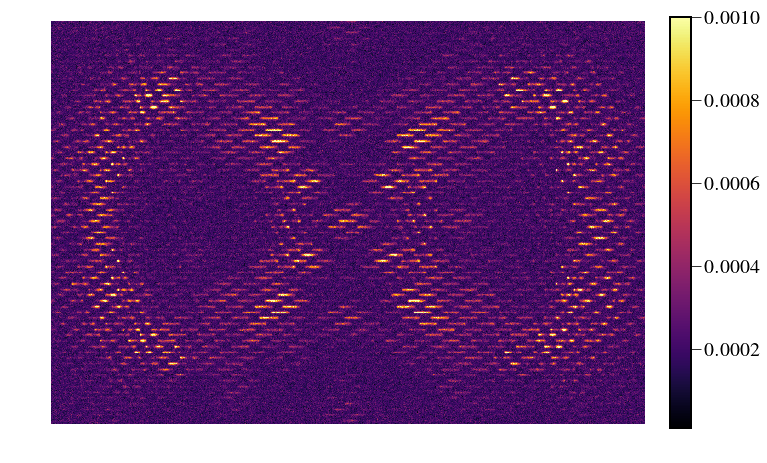
In [29]:
IFS_detector = Image(filename='/Users/mrizzo/IFS/OS5_SIM_2.0_t1000/average_47Umac/average_target.fits').data
plt.figure(figsize=(10,6))
img = IFS_detector[5,30:-30,30:-30]
print np.amin(img)
plt.imshow(img,cmap='inferno')
plt.colorbar(fraction=0.046, pad=0.04)
plt.axis('off')
plt.tight_layout()
plt.subplots_adjust(left=0.0,bottom=0)
# plt.savefig("/Users/mrizzo/Downloads/NoisyDetectorMap_Starshade.png",dpi=200)
crispy - INFO - Read data from HDU 1 of /Users/mrizzo/IFS/OS5_SIM_2.0_t1000/average_47Umac/average_target.fits
-2.66813e-09
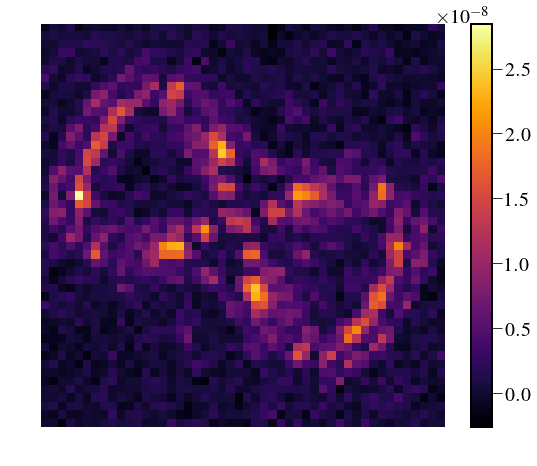
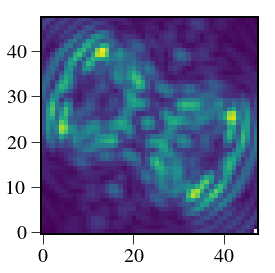
In [31]:
cube = Image(filename='/Users/mrizzo/IFS/OS5_SIM_2.0_noiseless/average/average_target_star_detector_red_optext_flatfielded.fits').data
print cube.shape
trim = 30
plt.imshow(np.sqrt(cube[15,trim:-trim,trim:-trim]))
print cube[15,trim:-trim,trim:-trim].shape
h,w = cube[15,trim:-trim,trim:-trim].shape
outarray = np.zeros((h,w*3))
outarray[:,:w] = cube[1,trim:-trim,trim:-trim]
outarray[:,w:w*2] = cube[11,trim:-trim,trim:-trim]
outarray[:,w*2:] = cube[-6,trim:-trim,trim:-trim]
plt.figure(figsize=(12,4))
plt.imshow(outarray,vmin=1e-10,norm=colors.PowerNorm(gamma=1./2.),cmap=cmap)
plt.axis('off')
plt.annotate('611 nm',xy=(w/2.-20,5),color='white',fontsize=26)
plt.annotate('660 nm',xy=(w+w/2.-20,5),color='white',fontsize=26)
plt.annotate('700 nm',xy=(2*w+w/2.-20,5),color='white',fontsize=26)
print np.amax(outarray)
plt.tight_layout()
plt.subplots_adjust(left=0.0, right=1, top=1, bottom=0)
plt.savefig("/Users/mrizzo/Downloads/OutputSlices.png",dpi=200)
crispy - INFO - Read data from HDU 1 of /Users/mrizzo/IFS/OS5_SIM_2.0_noiseless/average/average_target_star_detector_red_optext_flatfielded.fits
(19, 108, 108)
(48, 48)
3.31431770917e-08
Residuals¶
In [32]:
BB = Image(par.codeRoot+'/Inputs/BB.fits').data
BBres = Image(par.exportDir+'/BB_red_lstsq_resid.fits').data
# from crispy.params import Params
# par=Params(codefolder)
# par.wavecalDir = par.prefix+'/wavecalR50_660/'
# BB = Image(par.wavecalDir+'polychromeR50stack.fits').data
# BBres = reduceIFSMap(par,par.wavecalDir+'polychromeR50stack.fits',method='lstsq')
# BBres=BBres.data
# plt.figure(figsize=(20,12))
# plt.imshow(BBres)
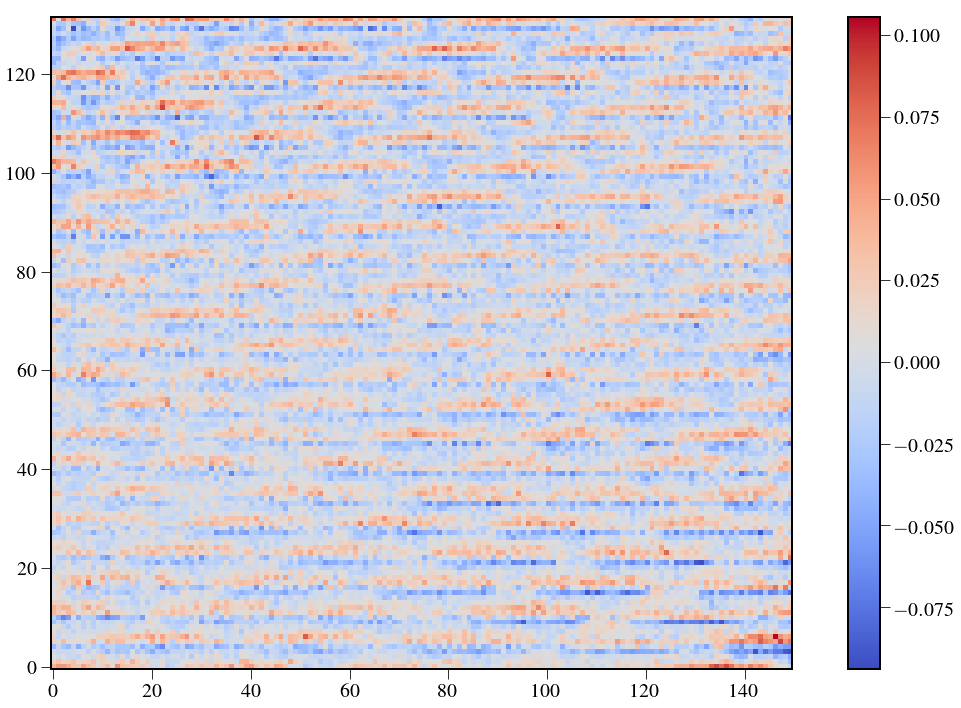
plt.figure(figsize=(20,12))
plt.imshow(BBres[850:-50,150:300]/np.amax(BB[850:-50,150:300]),cmap='coolwarm')
plt.colorbar()
# plt.imshow(BB[100:200,850:950])
# plt.imshow(BBres[100:200,850:950])
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
crispy - INFO - Read data from HDU 1 of ..//SimResults/BB_red_lstsq_resid.fits
Out[32]:
<matplotlib.colorbar.Colorbar at 0x1c1c4a7dd0>
In [34]:
from astropy.io import fits
BB = fits.getdata(par.codeRoot+"/Inputs/BB.fits")
BBres = fits.getdata(par.exportDir+'/BB_red_lstsq_resid.fits')
BBmod = fits.getdata(par.exportDir+'/BB_red_lstsq_model.fits')
cmap='viridis'
ftsize = 15
plt.figure(figsize=(10,10))
plt.subplot(131)
plt.imshow(BB[850:-100,150:300].T,vmin=-50,vmax=1000,cmap=cmap)
plt.axis('off')
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.title('Data',fontsize=ftsize)
plt.subplot(132)
plt.imshow(BBmod[850:-100,150:300].T,vmin=-50,vmax=1000,cmap=cmap)
plt.axis('off')
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.title('Model',fontsize=ftsize)
plt.subplot(133)
plt.imshow(100*BBres[850:-100,150:300].T/np.amax(BB[850:-120,150:300].T),vmin=-10,vmax=10,cmap=cmap)
plt.axis('off')
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.title('Residuals (\% of data max)',fontsize=ftsize)
plt.tight_layout()
plt.show()
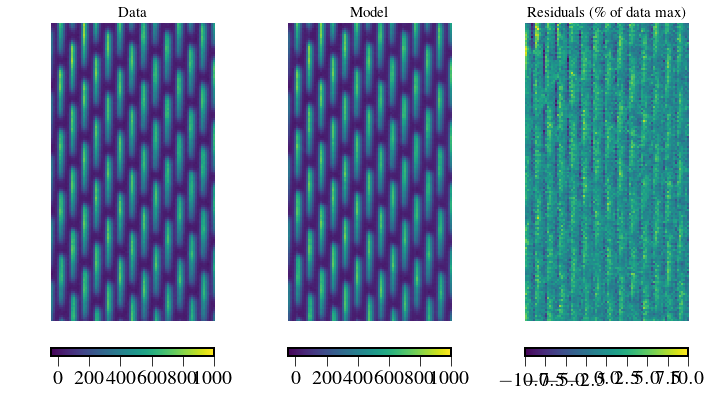
Simple broadband map¶
In [35]:
BB = Image(par.codeRoot+'/Inputs/BB.fits').data
flat = Image(par.codeRoot+'/Inputs/Flat637.fits').data
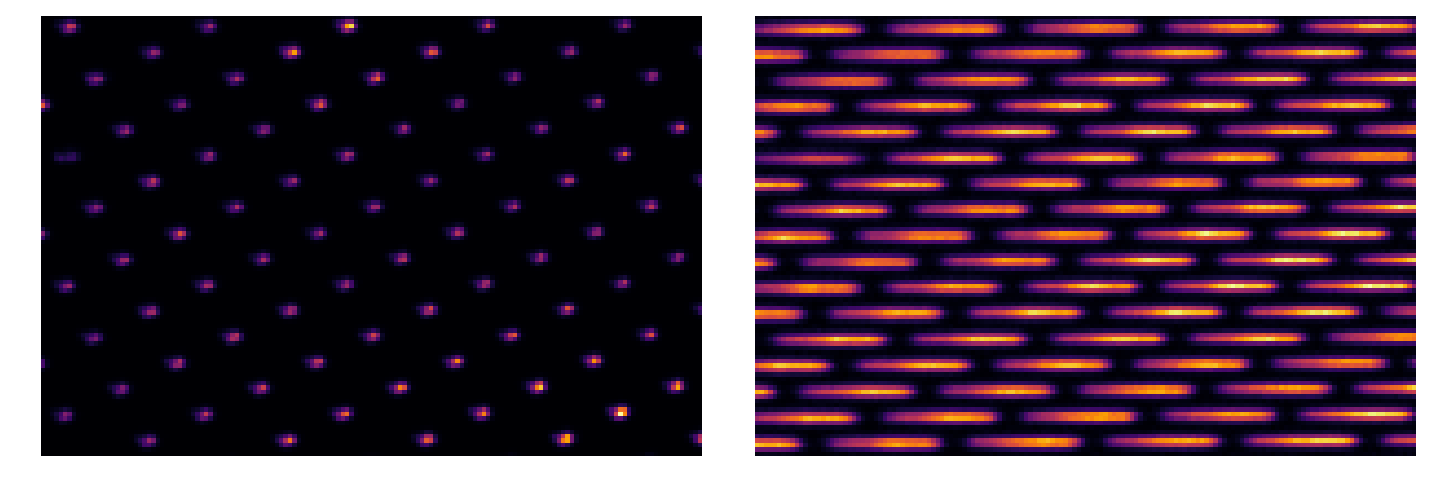
plt.figure(figsize=(20,12))
plt.subplot(121)
plt.imshow(flat[400:500,150:300],cmap='inferno')
plt.axis('off')
plt.subplot(122)
plt.imshow(BB[400:500,150:300],cmap='inferno')
plt.axis('off')
plt.tight_layout()
plt.savefig("/Users/mrizzo/Downloads/flatBB.png",dpi=200)
plt.show()
crispy - INFO - Read data from HDU 1 of ..//Inputs/BB.fits
crispy - INFO - Read data from HDU 1 of ..//Inputs/Flat637.fits
Bijan’s SPC PSF explanation¶
In [2]:
from crispy.tools.inputScene import adjust_krist_header
from crispy.IFS import polychromeIFS
os.chdir('/Users/mrizzo/IFS/crispy/crispy/WFIRST/')
from params import Params
par = Params()
par.hdr
offaxis = Image('/Users/mrizzo/IFS/OS5/offaxis/spc_offaxis_psf.fits')
adjust_krist_header(offaxis,lamc=660.)
fileshape = offaxis.data.shape
par.saveRotatedInput = True
lamlist = 660.*np.linspace(1.-par.BW/2.,1.+par.BW/2.,fileshape[0])
detector = polychromeIFS(par,lamlist,offaxis,QE=True)
crispy - INFO - Read data from HDU 0 of /Users/mrizzo/IFS/OS5/offaxis/spc_offaxis_psf.fits
crispy - INFO - The number of input pixels per lenslet is 5.000000
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/imagePlaneRot.fits
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Writing data to ..//SimResults/detectorFrame.fits
crispy - INFO - Done.
crispy - INFO - Performance: 37 seconds total
In [3]:
from astropy.io import fits
rotatedPSF = fits.getdata(par.exportDir+'/imagePlaneRot.fits')
fontsize=20
print(rotatedPSF.shape)
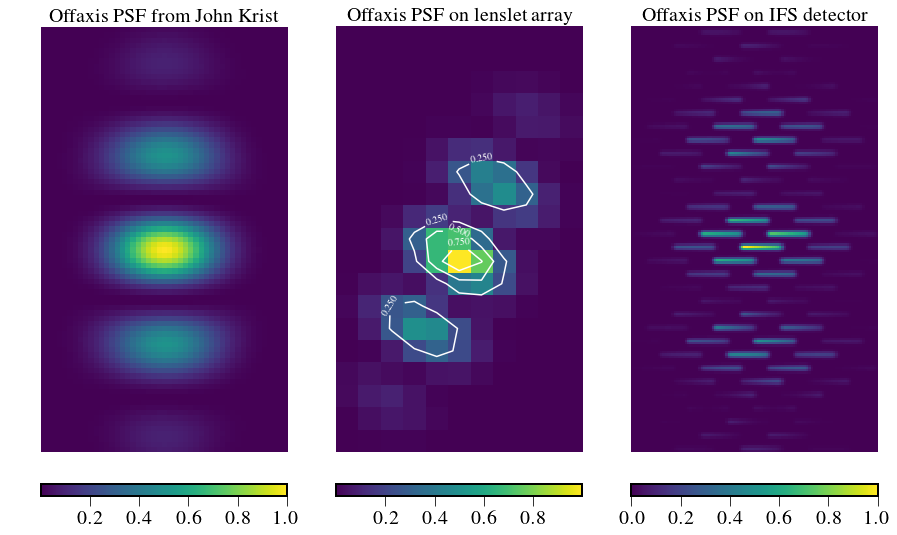
plt.figure(figsize=(15,15))
plt.subplot(132)
fig = rotatedPSF[25,21:40,43:54]
fig /= np.amax(fig)
plt.imshow(fig)
print fig[fig>0.5]
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.axis('off')
figshape = fig.shape
x = np.arange(figshape[1])
y = np.arange(figshape[0])
X,Y = np.meshgrid(x,y)
CS=plt.contour(X,Y,fig,levels=np.array([0.25,0.5,0.75]),colors='w')
plt.clabel(CS, inline=1, fontsize=10)
plt.title('Offaxis PSF on lenslet array',fontsize=fontsize)
plt.subplot(131)
fig = offaxis.data[25,90:171,175:222]
plt.imshow(fig/np.amax(fig))
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.title('Offaxis PSF from John Krist',fontsize=fontsize)
plt.axis('off')
plt.subplot(133)
fig = detector[410:600,640:750]
plt.imshow(fig/np.amax(fig))
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.axis('off')
plt.title('Offaxis PSF on IFS detector',fontsize=fontsize)
plt.show()
(45, 72, 72)
[ 0.66538638 1. 0.76297712 0.66954559 0.70729703]
In [4]:
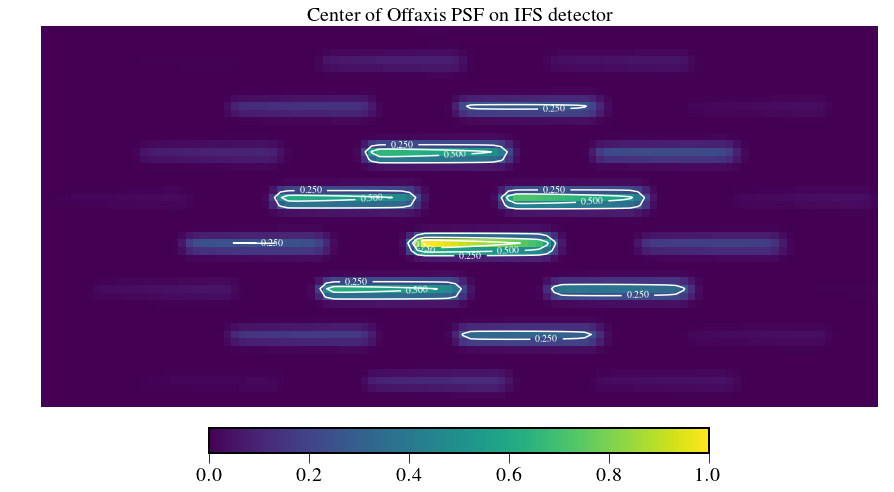
plt.figure(figsize=(15,10))
fig = detector[480:530,640:750]
fig /= np.amax(fig)
plt.imshow(fig)
plt.axis('off')
plt.title('Center of Offaxis PSF on IFS detector',fontsize=fontsize)
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
figshape=fig.shape
x = np.arange(figshape[1])
y = np.arange(figshape[0])
X,Y = np.meshgrid(x,y)
CS=plt.contour(X,Y,fig,levels=np.array([0.25,0.5,0.75]),colors='w')
plt.clabel(CS, inline=1, fontsize=10)
plt.show()