Introduction to crispy¶
This software is designed to simulate lenslet array-based Integral Field Spectrographs and their reduction process. This was developed within the context of NASA’s WFIRST Coronagraph mission.
In this notebook we go straight to the point and illustrate how to use the code. We require astropy, numpy. This notebook was written in a Python 3.5 kernel but should be backward compatible with Python 2.7.
Create a polychromatic flatfield¶
Initialization¶
First we need to load the various modules that we need. Since the notebook can be located anywhere, we need to add the location of the Python code to the path first.
In [1]:
import sys,os
import numpy as np
from crispy.tools.initLogger import getLogger
log = getLogger('crispy')
All the IFS parameters are contained within a Python class called ‘Params’. This class is initialized using only the path the ‘code’ directory from the repo. In particular, the Params class stores all the relevant paths. All parameters can be changed on the fly.
In [2]:
os.chdir('/Users/mrizzo/IFS/crispy/crispy/WFIRST/')
from params import Params
par = Params()
All the parameters are stored in this class. During the various steps of the software, they are appended to a header file.
In [3]:
par.hdr
Out[3]:
SIMPLE = T / conforms to FITS standard
BITPIX = 8 / array data type
NAXIS = 0 / number of array dimensions
EXTEND = T
COMMENT
COMMENT ************************************************************
COMMENT ********************** General parameters ******************
COMMENT ************************************************************
COMMENT
NLENS = 108 / # lenslets across array
PITCH = 0.000174 / Lenslet pitch (meters)
INTERLAC= 2.0 / Interlacing
PHILENS = 26.56505117707799 / Rotation angle of the lenslets (deg)
PIXSIZE = 1.3E-05 / Pixel size (meters)
LENSAMP = 0.5 / Lenslet sampling (lam/D)
LSAMPWAV= 660.0 / Lenslet sampling wavelength (nm)
FWHM = 2.0 / FHWM of PSFLet at detector (pixels)
FWHMLAM = 660.0 / Wavelength at which FWHM is defined (nm)
NPIX = 1024 / Number of detector pixels
BW = 0.18 / Bandwidth
PIXPRLAM= 2.0 / Pixels per resolution element
RESLSTSQ= 2.0 / Nspec per Nyq. sample for lstsq extraction
R = 50 / Spectral resolution
Create the flatfield¶
Creating a flatfield is a function built into the unitTests module. In this case, we will construct a cube of nlam wavelength slices, each having 512x512 pixels of value 1. The standard
In [4]:
from crispy.unitTests import testCreateFlatfield
help(testCreateFlatfield)
Help on function testCreateFlatfield in module crispy.unitTests:
testCreateFlatfield(par, pixsize=0.1, npix=512, pixval=1.0, Nspec=45, outname='flatfield.fits', useQE=True, method='optext')
Creates a polychromatic flatfield
Parameters
----------
par : Parameter instance
Contains all IFS parameters
pixsize: float
Pixel scale (lam/D)
npix: int
Each input frame has a pixel size npix x npix
pixval: float
Each input frame has a unform value pixval in photons per second per nm of bandwidth
Nspec: float
Optional input forcing the number of wavelengths bins used
outname: string
Name of flatfield image
useQE: boolean
Whether to take into account the wavelength-dependent QE of the detector
This test function will create a polychromatic flatfield at the wavelengths provided by the existing wavelength calibration. Those wavelengths can be retrieved as follows.
In [5]:
from crispy.tools.reduction import calculateWaveList
help(calculateWaveList)
lam_midpts,lam_endpts = calculateWaveList(par)
print(lam_midpts)
Help on function calculateWaveList in module crispy.tools.reduction:
calculateWaveList(par, lam_list=None, Nspec=None, method='lstsq')
Computes the wavelength lists corresponding to the center and endpoints of each
spectral bin. Wavelengths are separated by a constant value in log space. Number of
wavelengths depends on spectral resolution.
Parameters
----------
par: Parameter instance
Contains all IFS parameters
lam_list: list of floats
Usually this is left to None. If so, we use the wavelengths used for wavelength
calibration. Otherwise, we could decide to focus on a smaller/larger region of
the spectrum to retrieve. The final processed cubes will have bins centered
on lam_midpts
Nspec: int
If specified, forces the number of bins in the final cube (uses np.linspace)
Returns
-------
lam_midpts: list of floats
Wavelengths at the midpoint of each bin
lam_endpts: list of floats
Wavelengths at the edges of each bin
crispy - INFO - Reduced cube will have 19 wavelength bins
[ 603.48591977 609.29942699 615.16893696 621.09498916 627.07812827
633.11890422 639.21787224 645.37559291 651.5926322 657.86956154
664.20695787 670.60540367 677.06548705 683.58780178 690.17294734
696.821529 703.53415785 710.31145087 717.15403098]
Now let’s create our flatfield. By default it will be stored as par.unitTestsOutput/flatfield.fits
In [6]:
testCreateFlatfield(par,useQE=False)
crispy - INFO - Reduced cube will have 44 wavelength bins
crispy - INFO - The number of input pixels per lenslet is 5.016181
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming endpoints wavelist is given
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 52 seconds total
crispy - INFO - Writing data to ..//unitTestsOutputs/flatfield.fits
In [7]:
par.hdr
Out[7]:
SIMPLE = T / conforms to FITS standard
BITPIX = 8 / array data type
NAXIS = 0 / number of array dimensions
EXTEND = T
COMMENT
COMMENT ************************************************************
COMMENT ********************** General parameters ******************
COMMENT ************************************************************
COMMENT
NLENS = 108 / # lenslets across array
PITCH = 0.000174 / Lenslet pitch (meters)
INTERLAC= 2.0 / Interlacing
PHILENS = 26.56505117707799 / Rotation angle of the lenslets (deg)
PIXSIZE = 1.3E-05 / Pixel size (meters)
LENSAMP = 0.5 / Lenslet sampling (lam/D)
LSAMPWAV= 660.0 / Lenslet sampling wavelength (nm)
FWHM = 2.0 / FHWM of PSFLet at detector (pixels)
FWHMLAM = 660.0 / Wavelength at which FWHM is defined (nm)
NPIX = 1024 / Number of detector pixels
BW = 0.18 / Bandwidth
PIXPRLAM= 2.0 / Pixels per resolution element
RESLSTSQ= 2.0 / Nspec per Nyq. sample for lstsq extraction
R = 50 / Spectral resolution
COMMENT
COMMENT ************************************************************
COMMENT ********************** IFS Simulation ******************
COMMENT ************************************************************
COMMENT
SCALE = 5.016181205573144 / Factor by which the input slice is rescaled
INSLICES= 44 / Number of wavelengths in input cube
ADJUST = 0.98898 / Adjustment factor for rebinning error
Display results¶
Depending on your version of Python you might see a bunch of VisibleDeprecationWarning. We are working on fixing that. Let’s see what we got.
In [8]:
%pylab inline --no-import-all
matplotlib.rcParams['image.origin'] = 'lower'
matplotlib.rcParams['image.interpolation'] = 'nearest'
Populating the interactive namespace from numpy and matplotlib
In [9]:
from crispy.tools.image import Image
plt.figure(figsize=(10,10))
plt.imshow(Image(par.unitTestsOutputs+'/flatfield.fits').data,cmap='gray')
plt.colorbar()
crispy - INFO - Read data from HDU 1 of ..//unitTestsOutputs/flatfield.fits
Out[9]:
<matplotlib.colorbar.Colorbar at 0x117420a50>
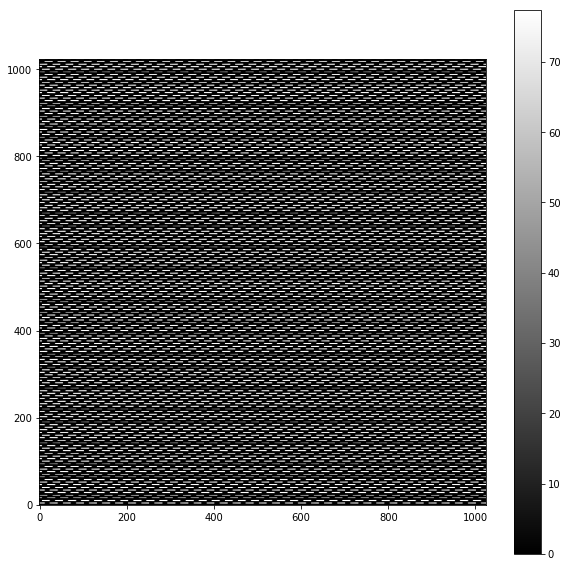
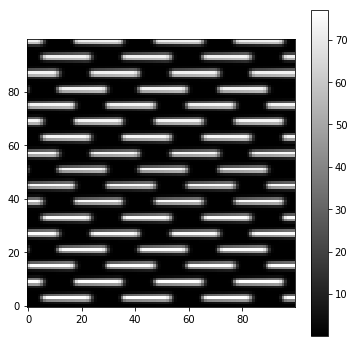
A zoom-in version is visible here:
In [10]:
plt.figure(figsize=(6,6))
img = Image(par.unitTestsOutputs+'/flatfield.fits').data
subsize = 50
plt.imshow(img[par.npix//2-subsize:par.npix//2+subsize,par.npix//2-subsize:par.npix//2+subsize],cmap='gray')
plt.colorbar()
crispy - INFO - Read data from HDU 1 of ..//unitTestsOutputs/flatfield.fits
Out[10]:
<matplotlib.colorbar.Colorbar at 0x117491210>
Simulate detector readout¶
We have also some routines that can add noise to the detector. Assuming that the input detector map is in photons per second, we can ‘detectorify’ this map by adding Poisson noise, read noise, CIC noise, and dark current noise. In the future, we will also implement Electron-multiplying noise and Traps.
In [11]:
from crispy.tools.image import Image
from crispy.tools.detector import readDetector,averageDetectorReadout
flat = Image(filename=par.unitTestsOutputs+'/flatfield.fits')
par.nonoise=False
# turn off photon counting otherwise things are strange
par.EMStats =False # turn off EM register statistics
par.PCmode = False # turn off photon counting threshold
read=readDetector(par,flat,inttime=100)
crispy - INFO - Read data from HDU 1 of ..//unitTestsOutputs/flatfield.fits
In [12]:
plt.figure(figsize=(6,6))
subsize = 15
plt.imshow(read[par.npix//2-subsize:par.npix//2+subsize,par.npix//2-subsize:par.npix//2+subsize],cmap='gray')
plt.colorbar()
Out[12]:
<matplotlib.colorbar.Colorbar at 0x1179236d0>

Let’s save the noisified frame to a new name. This is useful to show since writing to FITS is a very common task.
In [13]:
newImage = Image(data=read,header=par.hdr)
newImage.write(par.unitTestsOutputs+'/flatfield_noise.fits',clobber=True)
crispy - INFO - Writing data to ..//unitTestsOutputs/flatfield_noise.fits
Extraction¶
The reduction step is straightforward, as long as the wavelength calibration is good.
In [14]:
from crispy.IFS import reduceIFSMap
cube = reduceIFSMap(par,par.unitTestsOutputs+'/flatfield_noise.fits')
crispy - INFO - Read data from HDU 1 of ..//unitTestsOutputs/flatfield_noise.fits
crispy - INFO - Reduced cube will have 19 wavelength bins
crispy - INFO - Elapsed time: 1.748347s
Now we can display the cube interactively, or look it up with DS9 (it is located in par.exportDir)
In [15]:
import ipywidgets
def plt_ifs_optext(wchan):
plt.imshow(cube.data[wchan-1,:,:],
cmap='gist_heat')
plt.colorbar()
ipywidgets.interact(plt_ifs_optext, wchan=(1,cube.data.shape[0]));
Let’s look now at the header of the created file
In [16]:
cube.header
Out[16]:
SIMPLE = T / conforms to FITS standard
BITPIX = 8 / array data type
NAXIS = 0 / number of array dimensions
EXTEND = T
COMMENT
COMMENT ************************************************************
COMMENT ********************** General parameters ******************
COMMENT ************************************************************
COMMENT
NLENS = 108 / # lenslets across array
PITCH = 0.000174 / Lenslet pitch (meters)
INTERLAC= 2.0 / Interlacing
PHILENS = 26.56505117707799 / Rotation angle of the lenslets (deg)
PIXSIZE = 1.3E-05 / Pixel size (meters)
LENSAMP = 0.5 / Lenslet sampling (lam/D)
LSAMPWAV= 660.0 / Lenslet sampling wavelength (nm)
FWHM = 2.0 / FHWM of PSFLet at detector (pixels)
FWHMLAM = 660.0 / Wavelength at which FWHM is defined (nm)
NPIX = 1024 / Number of detector pixels
BW = 0.18 / Bandwidth
PIXPRLAM= 2.0 / Pixels per resolution element
RESLSTSQ= 2.0 / Nspec per Nyq. sample for lstsq extraction
R = 50 / Spectral resolution
COMMENT
COMMENT ************************************************************
COMMENT ********************** IFS Simulation ******************
COMMENT ************************************************************
COMMENT
SCALE = 5.016181205573144 / Factor by which the input slice is rescaled
INSLICES= 44 / Number of wavelengths in input cube
ADJUST = 0.98898 / Adjustment factor for rebinning error
COMMENT
COMMENT ************************************************************
COMMENT ********************** Detector readout ********************
COMMENT ************************************************************
COMMENT
TRANS = 1.0 / IFS Transmission factor
POL = 1.0 / Polarization losses
PHCTEFF = 1.0 / Photon counting efficiency
NONOISE = F / Ignore all noises?
POISSON = T / Poisson noise?
RN = 100.0 / Read noise (electrons/read)
CIC = 0.01 / Clock-induced charge
DARK = 0.0002 / Dark current
TRAPS = F / Use traps? T/F
EMSTATS = F / EM statistics?
EMGAIN = 2500.0 / Gain of the EM stage
PCBIAS = 200 / To make RN zero-mean
PCMODE = F / Photon counting mode?
LIFEFRAC= 0.0 / Mission life fraction (changes CTE if >0)
INTTIME = 100
COMMENT
COMMENT ************************************************************
COMMENT ********************** Cube Extraction *********************
COMMENT ************************************************************
COMMENT
R = 50 / Spectral resolution of final cube
CALDIR = 'wavecalR50_660' / Directory of wavelength solution
CUBEMODE= 'Optimal Extraction' / Method used to extract data cube
LAM_MIN = 603.4859197661123 / Minimum mid wavelength of extracted cube
LAM_MAX = 717.1540309800992 / Maximum mid wavelength of extracted cube
DLOGLAM = 0.009587107513934413 / Log spacing of extracted wavelength bins
NLAM = 19 / Number of extracted wavelengths
CTYPE1 = 'RA---TAN' / first parameter RA , projection TANgential
CTYPE2 = 'DEC--TAN' / second parameter DEC, projection TANgential
CRVAL1 = 0.0 / Reference X pixel value
CRVAL2 = 0.0 / Reference Y pixel value
CRPIX1 = 54 / Reference X pixel
CRPIX2 = 54 / Reference Y pixel
EQUINOX = 2000 / Equinox of coordinates
CD1_1 = -2.4845199749997E-06 / Rotation matrix coefficient
CD1_2 = 1.24225998749988E-06 / Rotation matrix coefficient
CD2_1 = 1.24225998749988E-06 / Rotation matrix coefficient
CD2_2 = 2.48451997499976E-06 / Rotation matrix coefficient
CTYPE3 = 'WAVE-LOG'
CUNIT3 = 'nm '
CRVAL3 = 603.4859197661123
CDELT3 = 6.307066216622808
CRPIX3 = 1
SMOOTHED= T / Cube smoothed over bad lenslets
Process cubes from John Krist¶
In [17]:
# cleans the header
par.makeHeader()
par.hdr
Out[17]:
SIMPLE = T / conforms to FITS standard
BITPIX = 8 / array data type
NAXIS = 0 / number of array dimensions
EXTEND = T
COMMENT
COMMENT ************************************************************
COMMENT ********************** General parameters ******************
COMMENT ************************************************************
COMMENT
NLENS = 108 / # lenslets across array
PITCH = 0.000174 / Lenslet pitch (meters)
INTERLAC= 2.0 / Interlacing
PHILENS = 26.56505117707799 / Rotation angle of the lenslets (deg)
PIXSIZE = 1.3E-05 / Pixel size (meters)
LENSAMP = 0.5 / Lenslet sampling (lam/D)
LSAMPWAV= 660.0 / Lenslet sampling wavelength (nm)
FWHM = 2.0 / FHWM of PSFLet at detector (pixels)
FWHMLAM = 660.0 / Wavelength at which FWHM is defined (nm)
NPIX = 1024 / Number of detector pixels
BW = 0.18 / Bandwidth
PIXPRLAM= 2.0 / Pixels per resolution element
RESLSTSQ= 2.0 / Nspec per Nyq. sample for lstsq extraction
R = 50 / Spectral resolution
In [18]:
# example of John Krist cube
offaxispsf= '/Users/mrizzo/IFS/OS5/offaxis/spc_offaxis_psf.fits'
offaxis = Image(offaxispsf)
offaxis.header
crispy - INFO - Read data from HDU 0 of /Users/mrizzo/IFS/OS5/offaxis/spc_offaxis_psf.fits
Out[18]:
SIMPLE = T / Written by IDL: Tue Feb 21 14:04:33 2017
BITPIX = -64 / Number of bits per data pixel
NAXIS = 3 / Number of data axes
NAXIS1 = 256 /Number of positions along axis 1
NAXIS2 = 256 /Number of positions along axis 2
NAXIS3 = 45 /Number of positions along axis 3
DATE = '2017-02-17' / Creation UTC (CCCC-MM-DD) date of FITS header
NLAM = 45 / Number of wavelengths
LAM_C = 0.800000 / Passband central wavelength in microns
LAM_MIN = 0.728000000000 / Minimum passband wavelength
LAM_MAX = 0.872000000000 / Maximum passband wavelength
PIXSIZE = 0.100000 / sampling in lam_c/D radians
OFFSET = 7.00000 / offset of star in lam_c/D radians
Shift the cube to the correct wavelength range corresponding to the band. We assume that the PSF doesn’t change, we just change the header keywords.
In [19]:
from crispy.tools.inputScene import adjust_krist_header
adjust_krist_header(offaxis,lamc=660.)
offaxis.header
Out[19]:
SIMPLE = T / Written by IDL: Tue Feb 21 14:04:33 2017
BITPIX = -64 / Number of bits per data pixel
NAXIS = 3 / Number of data axes
NAXIS1 = 256 /Number of positions along axis 1
NAXIS2 = 256 /Number of positions along axis 2
NAXIS3 = 45 /Number of positions along axis 3
DATE = '2017-02-17' / Creation UTC (CCCC-MM-DD) date of FITS header
NLAM = 45 / Number of wavelengths
LAM_C = 0.66 / Passband central wavelength in microns
LAM_MIN = 0.6005999999999999 / Minimum passband wavelength
LAM_MAX = 0.7193999999999999 / Maximum passband wavelength
PIXSIZE = 0.100000 / sampling in lam_c/D radians
OFFSET = 7.00000 / offset of star in lam_c/D radians
Process the cube¶
In [20]:
par.saveDetector=False
lamlist = np.linspace(offaxis.header['LAM_MIN'],offaxis.header['LAM_MAX'],offaxis.header['NLAM'])*1000
print(lamlist)
from crispy.IFS import polychromeIFS
detectorFrame = polychromeIFS(par,lamlist,offaxis,QE=True)
# save
Image(data=detectorFrame,header=par.hdr).write(par.exportDir+'/test_krist_cube.fits')
[ 600.6 603.3 606. 608.7 611.4 614.1 616.8 619.5 622.2 624.9
627.6 630.3 633. 635.7 638.4 641.1 643.8 646.5 649.2 651.9
654.6 657.3 660. 662.7 665.4 668.1 670.8 673.5 676.2 678.9
681.6 684.3 687. 689.7 692.4 695.1 697.8 700.5 703.2 705.9
708.6 711.3 714. 716.7 719.4]
crispy - INFO - The number of input pixels per lenslet is 5.000000
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 38 seconds total
crispy - INFO - Writing data to ..//SimResults/test_krist_cube.fits
Reduce the cube¶
In [21]:
reduced = reduceIFSMap(par,par.exportDir+'/test_krist_cube.fits')
crispy - INFO - Read data from HDU 1 of ..//SimResults/test_krist_cube.fits
crispy - INFO - Reduced cube will have 19 wavelength bins
crispy - INFO - Elapsed time: 1.641858s
In [22]:
def plt_ifs_optext(wchan):
plt.imshow(reduced.data[wchan-1,:,:],
cmap='gist_heat')
plt.colorbar()
ipywidgets.interact(plt_ifs_optext, wchan=(1,reduced.data.shape[0]));