Starting an IFS simulation from scratch¶
Let’s suppose that you want to model the ideal IFS and don’t have any idea (or don’t really care) about what the PSFLets look like and where they are located. In that case, you don’t need any datafile and can get started ASAP!
Let’s load things first.
In [1]:
import numpy as np
import glob
%pylab inline --no-import-all
plt.rc('font', family='serif', serif='Times',size=20)
plt.rc('text', usetex=True)
plt.rc('xtick', labelsize=20)
plt.rc('xtick.major', size=10)
plt.rc('ytick.major', size=10)
plt.rc('ytick', labelsize=20)
plt.rc('axes', labelsize=20)
plt.rc('figure',titlesize=20)
plt.rc('image',origin='lower',interpolation='nearest')
import sys
import os
Populating the interactive namespace from numpy and matplotlib
Initializes the logger used throughout the software¶
In [2]:
from crispy.tools.initLogger import getLogger
log = getLogger('crispy')
Operate in your working directory (where your parameter file lives)¶
In [3]:
os.chdir('/Users/mrizzo/IFS/crispy/crispy/WFIRST/')
Load parameters¶
In [4]:
from params import Params
par = Params()
par.hdr
Out[4]:
SIMPLE = T / conforms to FITS standard
BITPIX = 8 / array data type
NAXIS = 0 / number of array dimensions
EXTEND = T
COMMENT
COMMENT ************************************************************
COMMENT ********************** General parameters ******************
COMMENT ************************************************************
COMMENT
NLENS = 108 / # lenslets across array
PITCH = 0.000174 / Lenslet pitch (meters)
INTERLAC= 2.0 / Interlacing
PHILENS = 26.56505117707799 / Rotation angle of the lenslets (deg)
PIXSIZE = 1.3E-05 / Pixel size (meters)
LENSAMP = 0.5 / Lenslet sampling (lam/D)
LSAMPWAV= 660.0 / Lenslet sampling wavelength (nm)
FWHM = 2.0 / FHWM of PSFLet at detector (pixels)
FWHMLAM = 660.0 / Wavelength at which FWHM is defined (nm)
NPIX = 1024 / Number of detector pixels
BW = 0.18 / Bandwidth
PIXPRLAM= 2.0 / Pixels per resolution element
RESLSTSQ= 2.0 / Nspec per Nyq. sample for lstsq extraction
R = 50 / Spectral resolution
First off, you are going to trick crispy into building a fake wavelength calibration. This is usually important because in the real world, the wavelength calibration is the step that ensures that you can get good data out of your IFS images. For this exercise, you are going to use an a priori wavelength calibration (it’s built into crispy) to generate monochromatic flatfields. From these flatfields, you are going to build up your wavelength calibration - just like in real life!
Generate monochromatic flats¶
So, what’s our wavelength range? We are going to use the fiducial case of 600nm - 720nm, which is the first band of the WFIRST IFS. We are going to construct a few monochromatic flats across that range. The function we need is in the IFS module (that one has a lot of important functions).
In [5]:
from crispy.IFS import createWavecalFiles
help(createWavecalFiles)
Help on function createWavecalFiles in module crispy.IFS:
createWavecalFiles(par, lamlist, dlam=1.0)
Creates a set of monochromatic IFS images to be used in wavelength calibration step
Parameters
----------
par: Parameter instance
Contains all IFS parameters
lamlist: list or array of floats
List of discrete wavelengths at which to create a monochromatic flat
dlam: float
Width in nm of the small band for each of the monochromatic wavelengths.
Default is 1 nm. This has no effect unless we are trying to add any noise.
Notes
-----
This function populates the fields par.lamlist and par.filelist which are subsequently
used by the buildcalibrations function. If this createWavecalFiles is not called, the
two fields need to be populated manually with the set of files and wavelengths that
you want to use for the calibration.
So here we go:
In [6]:
lamc=660.
lamlist = np.arange(lamc*(1.-par.BW/2.),lamc*(1.+par.BW/2.)+5.,5.)
# lamlist = lamc*np.linspace(1.-BW/2.,1.+BW/2.,fileshape[0])
# let's create our new wavelength calibration directory to not mess up anything
# comment this out if this was already done and you are fine overriding the files in your folder
# try:
# os.makedirs(par.prefix+'/testWavecal660')
# except OSError:
# log.error('Wavecal folder already exists!')
# raise
log.info('Wavecal folder is: %s' % par.wavecalDir)
log.info(lamlist)
crispy - INFO - Wavecal folder is: ..//ReferenceFiles/wavecalR50_660/
crispy - INFO - [ 600.6 605.6 610.6 615.6 620.6 625.6 630.6 635.6 640.6 645.6
650.6 655.6 660.6 665.6 670.6 675.6 680.6 685.6 690.6 695.6
700.6 705.6 710.6 715.6 720.6]
In [7]:
createWavecalFiles(par,lamlist)
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_600.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_605.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_610.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_615.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_620.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_625.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_630.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_635.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_640.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_645.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_650.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_655.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_660.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 5 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_665.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_670.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_675.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_680.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_685.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 5 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_690.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_695.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 5 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_700.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_705.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_710.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_715.fits
crispy - INFO - The number of input pixels per lenslet is 4.995459
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming slices are evenly spread in wavelengths
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 4 seconds total
crispy - INFO - Writing data to ..//ReferenceFiles/wavecalR50_660/det_720.fits
Out[7]:
['..//ReferenceFiles/wavecalR50_660/det_600.fits',
'..//ReferenceFiles/wavecalR50_660/det_605.fits',
'..//ReferenceFiles/wavecalR50_660/det_610.fits',
'..//ReferenceFiles/wavecalR50_660/det_615.fits',
'..//ReferenceFiles/wavecalR50_660/det_620.fits',
'..//ReferenceFiles/wavecalR50_660/det_625.fits',
'..//ReferenceFiles/wavecalR50_660/det_630.fits',
'..//ReferenceFiles/wavecalR50_660/det_635.fits',
'..//ReferenceFiles/wavecalR50_660/det_640.fits',
'..//ReferenceFiles/wavecalR50_660/det_645.fits',
'..//ReferenceFiles/wavecalR50_660/det_650.fits',
'..//ReferenceFiles/wavecalR50_660/det_655.fits',
'..//ReferenceFiles/wavecalR50_660/det_660.fits',
'..//ReferenceFiles/wavecalR50_660/det_665.fits',
'..//ReferenceFiles/wavecalR50_660/det_670.fits',
'..//ReferenceFiles/wavecalR50_660/det_675.fits',
'..//ReferenceFiles/wavecalR50_660/det_680.fits',
'..//ReferenceFiles/wavecalR50_660/det_685.fits',
'..//ReferenceFiles/wavecalR50_660/det_690.fits',
'..//ReferenceFiles/wavecalR50_660/det_695.fits',
'..//ReferenceFiles/wavecalR50_660/det_700.fits',
'..//ReferenceFiles/wavecalR50_660/det_705.fits',
'..//ReferenceFiles/wavecalR50_660/det_710.fits',
'..//ReferenceFiles/wavecalR50_660/det_715.fits',
'..//ReferenceFiles/wavecalR50_660/det_720.fits']
In [8]:
par.R = 50
# par.wavecalDir = ''
Let’s take a look at what we did.
In [9]:
listfiles = glob.glob(par.wavecalDir+'*.fits')
print listfiles
['..//ReferenceFiles/wavecalR50_660/det_600.fits', '..//ReferenceFiles/wavecalR50_660/det_605.fits', '..//ReferenceFiles/wavecalR50_660/det_610.fits', '..//ReferenceFiles/wavecalR50_660/det_615.fits', '..//ReferenceFiles/wavecalR50_660/det_620.fits', '..//ReferenceFiles/wavecalR50_660/det_625.fits', '..//ReferenceFiles/wavecalR50_660/det_630.fits', '..//ReferenceFiles/wavecalR50_660/det_635.fits', '..//ReferenceFiles/wavecalR50_660/det_640.fits', '..//ReferenceFiles/wavecalR50_660/det_645.fits', '..//ReferenceFiles/wavecalR50_660/det_650.fits', '..//ReferenceFiles/wavecalR50_660/det_655.fits', '..//ReferenceFiles/wavecalR50_660/det_660.fits', '..//ReferenceFiles/wavecalR50_660/det_665.fits', '..//ReferenceFiles/wavecalR50_660/det_670.fits', '..//ReferenceFiles/wavecalR50_660/det_675.fits', '..//ReferenceFiles/wavecalR50_660/det_680.fits', '..//ReferenceFiles/wavecalR50_660/det_685.fits', '..//ReferenceFiles/wavecalR50_660/det_690.fits', '..//ReferenceFiles/wavecalR50_660/det_695.fits', '..//ReferenceFiles/wavecalR50_660/det_700.fits', '..//ReferenceFiles/wavecalR50_660/det_705.fits', '..//ReferenceFiles/wavecalR50_660/det_710.fits', '..//ReferenceFiles/wavecalR50_660/det_715.fits', '..//ReferenceFiles/wavecalR50_660/det_720.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam600.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam605.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam610.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam615.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam620.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam625.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam630.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam635.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam640.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam645.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam650.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam655.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam660.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam665.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam670.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam675.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam680.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam685.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam690.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam695.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam700.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam705.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam710.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam715.fits', '..//ReferenceFiles/wavecalR50_660/hires_psflets_lam720.fits', '..//ReferenceFiles/wavecalR50_660/polychromekeyR50.fits', '..//ReferenceFiles/wavecalR50_660/polychromeR50.fits', '..//ReferenceFiles/wavecalR50_660/PSFloc.fits', '..//ReferenceFiles/wavecalR50_660/PSFwidths.fits']
Now let’s show some images. We use the Image module of crispy for convenience.
In [10]:
from crispy.tools.image import Image
plt.figure(figsize=(10,10))
plt.imshow(Image(listfiles[0]).data)
plt.colorbar(fraction=0.046, pad=0.04)
plt.show()
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_600.fits
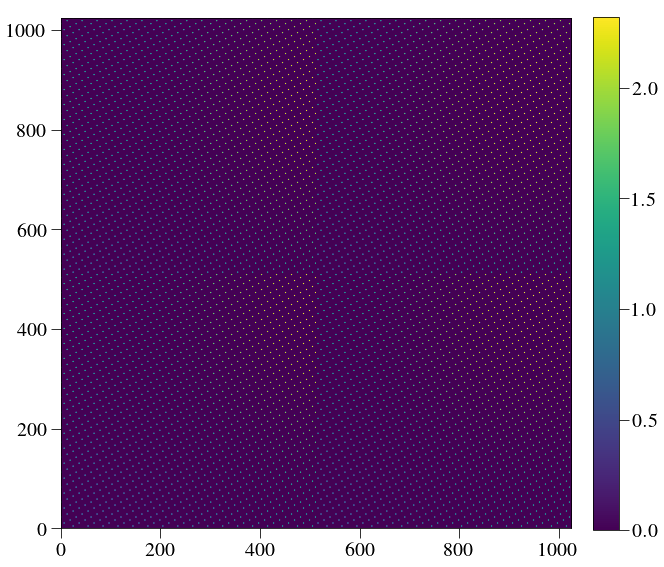
And a little zoom in:
In [11]:
plt.figure(figsize=(10,10))
plt.imshow(Image(listfiles[0]).data[450:550,450:550])
plt.colorbar(fraction=0.046, pad=0.04)
plt.show()
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_600.fits
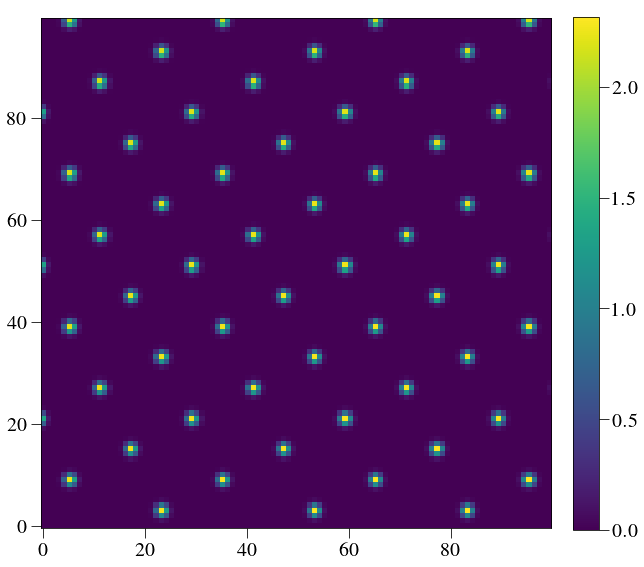
The PSFLets are slowly changing because the geometry that we chose results in almost integer spacing in both X and Y between neighboring lenslets. If you want more diversity (usually it is preferable to have diversity), then change the lenslet size of detector pitch by a little bit.
Run a complete wavelength calibration¶
Now that we have monochromatic flats, it is time to run a wavelength calibration, just like one would do in any IFS. The function we need is in crispy.tools.wavecal. This is a much more complicated function.
In [12]:
from crispy.tools.wavecal import buildcalibrations
help(buildcalibrations)
Help on function buildcalibrations in module crispy.tools.wavecal:
buildcalibrations(par, filelist=None, lamlist=None, order=3, inspect=False, genwavelengthsol=False, makehiresPSFlets=False, makePolychrome=False, makehiresPolychrome=False, makePSFWidths=False, savehiresimages=True, borderpix=4, upsample=5, nsubarr=3, parallel=True, inspect_first=True, apodize=False, lamsol=None, threshold=0.0)
Master wavelength calibration function
Parameters
----------
par : Parameter instance
Contains all IFS parameters
filelist: list of strings (optional)
List of the fits files that contain the monochromatic calibration files. If None (default),
use the files in par.filelist
lamlist: list of floats (optional)
Wavelengths in nm at which the files are taken. If None (default),
use the files in par.lamlist
order: int
Order of the polynomial used to fit the PSFLet positions across the detector
genwavelengthsol: Boolean
If True, generate the wavelength calibration. Creates a text file with all
polynomial coefficients that best fit the PSFLet positions at each wavelength.
If False, then load an already-generated file.
inspect: Boolean
Whether or not to create PNG files that overlay PSFLet fitted position on the
monochromatic pictures, to visually inspect the fitting results
inspect_first: Boolean
Whether or not to create a PNG file that overlays PSFLet fitted position on the
monochromatic picture of the first file, to visually inspect the fitting results
makehiresPSFlets: Boolean
Whether or not to do a high-resolution fitting of the PSFs, using the sampling
diversity. This requires high-SNR monochromatic images.
makePolychrome: Boolean
Whether or not to build the polychrome cube used in the least squares extraction
makePSFWidths: Boolean
Whether or not to fit the PSFLet widths using the high-res PSFLets
makehiresPolychrome: Boolean
Whether or not to build a polychrome cube at a high spatial resolution for future
subpixel interpolations
savehiresimages: Boolean
Whether to save fits files with the high-res PSFLets
borderpix: int
Number of pixels that are not taken into account towards the edges of the detector
upsample: int
Upsampling factor for each high-resolution PSFLet
nsubarr: int
Detector will be divided into nsubarr x nsubarr regions. A high-resolution PSFLet
will be determined in each region from the average of all PSFLets within that
region
parallel: Boolean
Whether or not to parallelize the computation for the high-resolution PSFLet and
polychrome computation. The wavelength calibration step cannot be parallelized since
each wavelength uses the previous wavelength solution as a guess input.
apodize: Boolean
Whether to fit the spots only using lenslets within a circle, ignoring the corners of
the detector
lamsol: 2D array
Optional argument that, if not None and if genwavelengthsol==False, will take the argument
and use it as the current wavelength calibration to build the polychrome.
threshold: float
Threshold under which to zero out the polychrome. This is only useful for reducing
the file size of the polychrome, and has only very little impact on the extraction.
To be safe, for science extractions threshold should be kept at its default value of 0.0
Notes
-----
This function generates all the files required to process IFS cubes:
lamsol.dat: contains a list of the wavelengths and the polynomial coefficients that
describe the X,Y positions of all lenslets on the detector as a function
of lenslet position on the lenslet array.
polychromekeyRXX.fits: where XX is replaced by the spectral resolution defined in the
parameters file. This is a multi-extension fits file with:
- a list of the central wavelengths at which the final cube will be reduced to
- an array of the X positions of all lenslets
- an array of the Y positions of all lenslets
- an array of booleans indicating whether that lenslet is good or not
(e.g. when it is outside of the detector area)
polychromeRXX.fits: 3D arrays of size Nspec x Npix x Npix with maps of the PSFLets put in their correct
positions for each wavelength bins that we want in the output cube. Each PSFLet
in each wavelength slice is used for least-squares fitting.
hiresPolychromeRXX.fits: same as polychromeRXX.fits but this time using the high-resolution PSFLets
PSFLoc.fits: nsubarr x nsubarr array of 2D high-resolution PSFLets at each location
in the detector.
In [13]:
# we can check that the file list has been loaded:
log.info('Wavecal files: %s' % par.filelist)
crispy - INFO - Wavecal files: ['..//ReferenceFiles/wavecalR50_660/det_600.fits', '..//ReferenceFiles/wavecalR50_660/det_605.fits', '..//ReferenceFiles/wavecalR50_660/det_610.fits', '..//ReferenceFiles/wavecalR50_660/det_615.fits', '..//ReferenceFiles/wavecalR50_660/det_620.fits', '..//ReferenceFiles/wavecalR50_660/det_625.fits', '..//ReferenceFiles/wavecalR50_660/det_630.fits', '..//ReferenceFiles/wavecalR50_660/det_635.fits', '..//ReferenceFiles/wavecalR50_660/det_640.fits', '..//ReferenceFiles/wavecalR50_660/det_645.fits', '..//ReferenceFiles/wavecalR50_660/det_650.fits', '..//ReferenceFiles/wavecalR50_660/det_655.fits', '..//ReferenceFiles/wavecalR50_660/det_660.fits', '..//ReferenceFiles/wavecalR50_660/det_665.fits', '..//ReferenceFiles/wavecalR50_660/det_670.fits', '..//ReferenceFiles/wavecalR50_660/det_675.fits', '..//ReferenceFiles/wavecalR50_660/det_680.fits', '..//ReferenceFiles/wavecalR50_660/det_685.fits', '..//ReferenceFiles/wavecalR50_660/det_690.fits', '..//ReferenceFiles/wavecalR50_660/det_695.fits', '..//ReferenceFiles/wavecalR50_660/det_700.fits', '..//ReferenceFiles/wavecalR50_660/det_705.fits', '..//ReferenceFiles/wavecalR50_660/det_710.fits', '..//ReferenceFiles/wavecalR50_660/det_715.fits', '..//ReferenceFiles/wavecalR50_660/det_720.fits']
One important aspect is that we can choose to use the same models for PSFLets as the one that was used to construct the monochromatic flatfields. This basically assumes that we have 100% knowledge of all the PSFLets, and can be set if par.gaussian_hires=True. Otherwise, we can actually use the deconvolution functions within crispy to approximate the lenslet PSFLets, by dividing up the detector into nsubarr^2 regions and averaging the PSFLets over these regions. Let’s assume for now that we have excellent models of our PSFLets.
In [14]:
par.gaussian_hires=True # assuming we know the PSFLets perfectly a priori
If you choose to change that to False, you should also change the following nsubarr parameter to >4 since there is likely variation of the PSFLet across the field of view. Let’s go! This takes a few minutes. It will finish by a parallel process so don’t worry if your computer heats up a bit.
In [15]:
buildcalibrations(par,
inspect=False, # if True, constructs a bunch of images to verify a good calibration
genwavelengthsol=True, # Compute wavelength at the center of all pixels
makehiresPSFlets=True, # this requires very high SNR on the monochromatic frames
makePSFWidths=True,
makePolychrome=True, # This is needed to use least squares extraction
upsample=3, # upsampling factor of the high-resolution PSFLets
nsubarr=3, # the detector is divided into nsubarr^2 regions for PSFLet averaging
apodize=False, # to match PSFlet spot locations, only use the inner circular part of the
#detector, hence discarding the corners of the detector where lenslets are
#distorted
threshold=1e-4,
)
crispy - INFO - Building calibration files, placing results in ..//ReferenceFiles/wavecalR50_660/
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_600.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_600.fits
crispy - INFO - Initializing PSFlet location transformation coefficients
crispy - INFO - Performing initial optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_600.fits
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_600.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_605.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_605.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_610.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_610.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_615.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_615.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_620.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_620.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_625.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_625.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_630.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_630.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_635.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_635.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_640.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_640.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_645.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_645.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_650.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_650.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_655.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_655.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_660.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_660.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_665.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_665.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_670.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_670.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_675.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_675.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_680.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_680.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_685.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_685.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_690.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_690.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_695.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_695.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_700.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_700.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_705.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_705.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_710.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_710.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_715.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_715.fits
crispy - INFO - Read data from HDU 1 of ..//ReferenceFiles/wavecalR50_660/det_720.fits
crispy - INFO - Initializing transformation coefficients with previous values
crispy - INFO - Performing final optimization of PSFlet location transformation coefficients for frame ..//ReferenceFiles/wavecalR50_660/det_720.fits
crispy - INFO - Saving wavelength solution to ..//ReferenceFiles/wavecalR50_660/lamsol.dat
crispy - INFO - Computing wavelength values at pixel centers
crispy - INFO - Making high-resolution PSFLet models
crispy - INFO - Starting parallel computation
crispy - INFO - Computing PSFLet widths...
crispy - INFO - Reduced cube will have 19 wavelength bins
crispy - INFO - Making polychrome cube
crispy - INFO - Saving polychrome cube
crispy - INFO - Saving wavelength calibration cube
crispy - INFO - Total time elapsed: 264 s
Let’s take a look at the files we constructed:
In [16]:
os.listdir(par.wavecalDir)
Out[16]:
['.DS_Store',
'det_600.fits',
'det_605.fits',
'det_610.fits',
'det_615.fits',
'det_620.fits',
'det_625.fits',
'det_630.fits',
'det_635.fits',
'det_640.fits',
'det_645.fits',
'det_650.fits',
'det_655.fits',
'det_660.fits',
'det_665.fits',
'det_670.fits',
'det_675.fits',
'det_680.fits',
'det_685.fits',
'det_690.fits',
'det_695.fits',
'det_700.fits',
'det_705.fits',
'det_710.fits',
'det_715.fits',
'det_720.fits',
'hires_psflets_lam600.fits',
'hires_psflets_lam605.fits',
'hires_psflets_lam610.fits',
'hires_psflets_lam615.fits',
'hires_psflets_lam620.fits',
'hires_psflets_lam625.fits',
'hires_psflets_lam630.fits',
'hires_psflets_lam635.fits',
'hires_psflets_lam640.fits',
'hires_psflets_lam645.fits',
'hires_psflets_lam650.fits',
'hires_psflets_lam655.fits',
'hires_psflets_lam660.fits',
'hires_psflets_lam665.fits',
'hires_psflets_lam670.fits',
'hires_psflets_lam675.fits',
'hires_psflets_lam680.fits',
'hires_psflets_lam685.fits',
'hires_psflets_lam690.fits',
'hires_psflets_lam695.fits',
'hires_psflets_lam700.fits',
'hires_psflets_lam705.fits',
'hires_psflets_lam710.fits',
'hires_psflets_lam715.fits',
'hires_psflets_lam720.fits',
'inspection_600.png',
'lamsol.dat',
'polychromekeyR50.fits',
'polychromeR50.fits',
'polychromeR50.fits.gz',
'polychromeR50stack.fits.gz',
'PSFloc.fits',
'PSFwidths.fits']
Some of these files are described and explained in the papers associated with this work.
Construct fields to extract¶
Now, let’s construct a field that we will want to extract. For example, we will use a flatfield.
In [17]:
from crispy.unitTests import testCreateFlatfield
help(testCreateFlatfield)
Help on function testCreateFlatfield in module crispy.unitTests:
testCreateFlatfield(par, pixsize=0.1, npix=512, pixval=1.0, Nspec=45, outname='flatfield.fits', useQE=True, method='optext')
Creates a polychromatic flatfield
Parameters
----------
par : Parameter instance
Contains all IFS parameters
pixsize: float
Pixel scale (lam/D)
npix: int
Each input frame has a pixel size npix x npix
pixval: float
Each input frame has a unform value pixval in photons per second per nm of bandwidth
Nspec: float
Optional input forcing the number of wavelengths bins used
outname: string
Name of flatfield image
useQE: boolean
Whether to take into account the wavelength-dependent QE of the detector
In [18]:
par.savePoly=True
par.saveRotatedInput = True
testCreateFlatfield(par,useQE=False)
crispy - INFO - Reduced cube will have 44 wavelength bins
crispy - INFO - The number of input pixels per lenslet is 5.016181
crispy - INFO - Using PSFlet gaussian approximation
crispy - WARNING - Assuming endpoints wavelist is given
crispy - INFO - Creating Gaussian PSFLet templates
crispy - INFO - Writing data to ..//SimResults/imagePlaneRot.fits
crispy - INFO - Writing data to ..//SimResults/detectorFramepoly.fits
crispy - INFO - Done.
crispy - INFO - Performance: 50 seconds total
crispy - INFO - Writing data to ..//unitTestsOutputs/flatfield.fits
In [19]:
plt.figure(figsize=(10,10))
plt.imshow(Image(par.unitTestsOutputs+'/flatfield.fits').data)
plt.show()
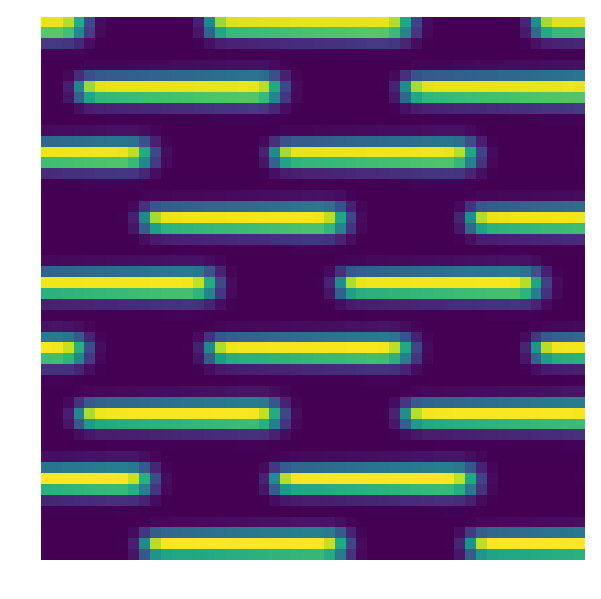
plt.figure(figsize=(10,10))
plt.imshow(Image(par.unitTestsOutputs+'/flatfield.fits').data[500:550,500:550])
plt.axis('off')
plt.show()
crispy - INFO - Read data from HDU 1 of ..//unitTestsOutputs/flatfield.fits
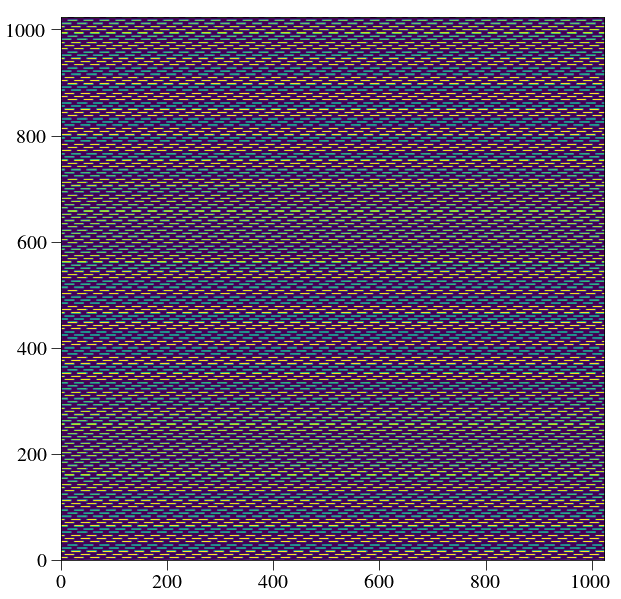
crispy - INFO - Read data from HDU 1 of ..//unitTestsOutputs/flatfield.fits
Reduce the flatfield!¶
In [20]:
from crispy.IFS import reduceIFSMap
help(reduceIFSMap)
detector = reduceIFSMap(par,par.unitTestsOutputs+'/flatfield.fits',method='lstsq_conv',smoothbad=False,fitbkgnd=False,niter=1)
Help on function reduceIFSMap in module crispy.IFS:
reduceIFSMap(par, IFSimageName, method='optext', smoothbad=True, name=None, hires=False, dy=3, fitbkgnd=True, specialPolychrome=None, returnall=False, niter=10, pixnoise=0.0, normpsflets=False)
Main reduction function
Uses various routines to extract an IFS detector map into a spectral-spatial cube.
Parameters
----------
par : Parameter instance
Contains all IFS parameters
IFSimageName : string or 2D ndarray
Path of image file, of 2D ndarray.
method : 'lstsq', 'optext'
Method used for reduction.
'lstsq': use the knowledge of the PSFs at each location and each wavelength and fits
the microspectrum as a weighted sum of these PSFs in the least-square sense. Can weigh the data by its variance.
'optext': use a matched filter to appropriately weigh each pixel and assign the fluxes, making use of the inverse
wavlength calibration map. Then remap each microspectrum onto the desired wavelengths
Returns
-------
cube: 3D ndarray
Reduced IFS cube
crispy - INFO - Read data from HDU 1 of ..//unitTestsOutputs/flatfield.fits
crispy - INFO - Reduced cube will have 19 wavelength bins
crispy - INFO - Writing data to ..//SimResults/flatfield_red_lstsq_conv_resid.fits
crispy - INFO - Writing data to ..//SimResults/flatfield_red_lstsq_conv_model.fits
crispy - INFO - Elapsed time: 11.791295s
Let’s plot what we have got
In [21]:
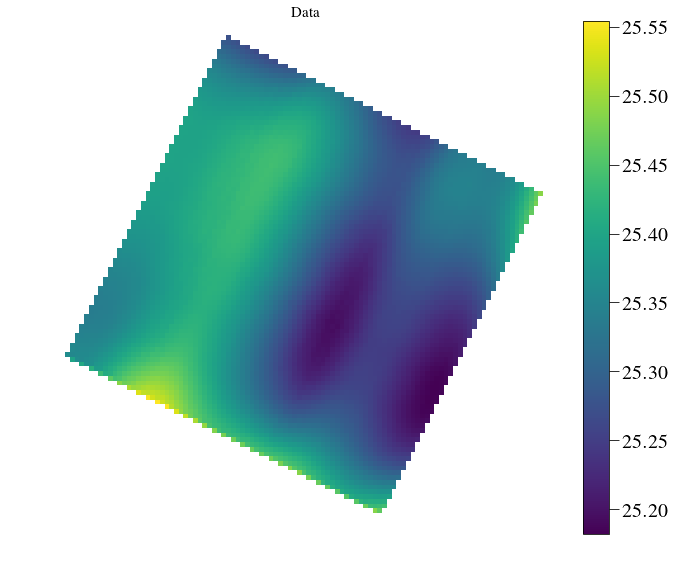
ftsize = 15
cmap='viridis'
plt.figure(figsize=(10,10))
plt.imshow(Image(par.exportDir+'/flatfield_red_lstsq.fits').data[5],cmap=cmap)
plt.axis('off')
plt.colorbar(fraction=0.046, pad=0.04)
plt.title('Data',fontsize=ftsize)
plt.show()
crispy - INFO - Read data from HDU 1 of ..//SimResults/flatfield_red_lstsq.fits
crispy - INFO - Read inverse variance from HDU 2 of ..//SimResults/flatfield_red_lstsq.fits
In [22]:
from astropy.io import fits
BB = fits.getdata(par.unitTestsOutputs+"/flatfield.fits")
BBres = fits.getdata(par.exportDir+'/flatfield_red_lstsq_resid.fits')
BBmod = fits.getdata(par.exportDir+'/flatfield_red_lstsq_model.fits')
plt.figure(figsize=(10,10))
plt.subplot(131)
plt.imshow(BB[850:-100,150:300].T,vmin=0,vmax=np.amax(BB[850:-100,150:300]),cmap=cmap)
plt.axis('off')
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.title('Data',fontsize=ftsize)
plt.subplot(132)
plt.imshow(BBmod[850:-100,150:300].T,vmin=0,vmax=np.amax(BB[850:-100,150:300]),cmap=cmap)
plt.axis('off')
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.title('Model',fontsize=ftsize)
plt.subplot(133)
plt.imshow(100*BBres[850:-100,150:300].T/np.amax(BB[850:-120,150:300].T),vmin=-2,vmax=2,cmap=cmap)
plt.axis('off')
plt.colorbar(fraction=0.046, pad=0.04,orientation='horizontal')
plt.title('Residuals (\% of data max)',fontsize=ftsize)
plt.tight_layout()
plt.show()
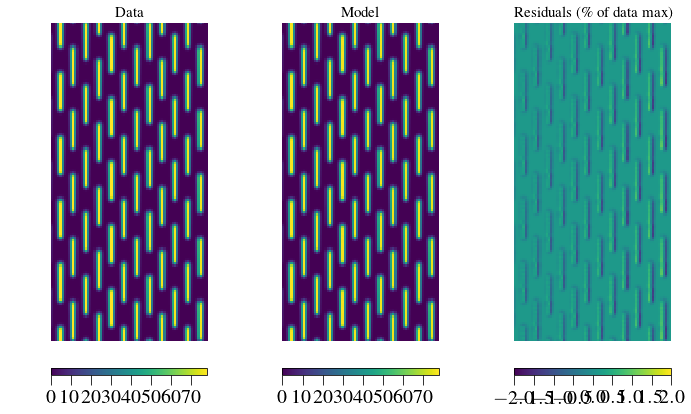
The residuals are caused by an imperfect wavelength calibration (small errors in centroids). In this region it is less than +/-2% of the peak data value, and the induced errors in recovered flux are +/-1% in slice number 5 (see above). These are systematic errors that we can calibrate by using an IFS flat field.
If we want to know the correspondence between slice and wavelength, we can do the following
In [23]:
from crispy.tools.reduction import calculateWaveList
lam_midpts,lam_endpts = calculateWaveList(par,method='lstsq')
print('Wavelengths at midpoints for lstsq (nm): ',lam_midpts)
print('Wavelengths at endpoints for lstsq (nm): ',lam_endpts)
lam_midpts,lam_endpts = calculateWaveList(par,method='optext')
print('Wavelengths at midpoints for optext (nm): ',lam_midpts)
print('Wavelengths at endpoints for optext (nm): ',lam_endpts)
crispy - INFO - Reduced cube will have 19 wavelength bins
('Wavelengths at midpoints for lstsq (nm): ', array([ 603.48591977, 609.29942699, 615.16893696, 621.09498916,
627.07812827, 633.11890422, 639.21787224, 645.37559291,
651.5926322 , 657.86956154, 664.20695787, 670.60540367,
677.06548705, 683.58780178, 690.17294734, 696.821529 ,
703.53415785, 710.31145087, 717.15403098]))
('Wavelengths at endpoints for lstsq (nm): ', array([ 600.6 , 606.38570655, 612.22714804, 618.12486136,
624.0793886 , 630.09127706, 636.16107931, 642.28935325,
648.47666214, 654.72357468, 661.03066505, 667.39851296,
673.82770368, 680.31882816, 686.87248301, 693.4892706 ,
700.1697991 , 706.91468254, 713.72454087, 720.6 ]))
crispy - INFO - Reduced cube will have 19 wavelength bins
('Wavelengths at midpoints for optext (nm): ', array([ 603.48591977, 609.29942699, 615.16893696, 621.09498916,
627.07812827, 633.11890422, 639.21787224, 645.37559291,
651.5926322 , 657.86956154, 664.20695787, 670.60540367,
677.06548705, 683.58780178, 690.17294734, 696.821529 ,
703.53415785, 710.31145087, 717.15403098]))
('Wavelengths at endpoints for optext (nm): ', array([ 600.6 , 606.38570655, 612.22714804, 618.12486136,
624.0793886 , 630.09127706, 636.16107931, 642.28935325,
648.47666214, 654.72357468, 661.03066505, 667.39851296,
673.82770368, 680.31882816, 686.87248301, 693.4892706 ,
700.1697991 , 706.91468254, 713.72454087, 720.6 ]))
Start from known wavecal¶
In the case where we want to use another wavecal (e.g. obtained from lab measurements), we can set this up with par.PSFLetPositions = True
In [24]:
# par.PSFLetPositions = True
# par.wavecalDir = par.prefix+'/Calibra_170425/'
# watch out to be sure you have the correct R that matches the wavecal! In this case, it is R=70
#par.R = 70
Repeat the flatfield exercise
In [25]:
#from crispy.unitTests import testCreateFlatfield
#testCreateFlatfield(par,useQE=False)
In [26]:
# par.PSFLetPositions = False
# par.R=50
# par.wavecalDir = par.prefix+'/wavecalR50_SC/'
In [27]:
# from crispy.unitTests import testCrosstalk
# testCrosstalk(par,pixval=25,method='optext',useQE=False)
# extracted = reduceIFSMap(par,par.unitTestsOutputs+'/crosstalk.fits',method='lstsq',smoothbad=False)
In [28]:
# extracted = reduceIFSMap(par,par.unitTestsOutputs+'/flatfield.fits',method='lstsq',smoothbad=False)